Tularemia is a disease caused by
Francisella tularensis, a highly pathogenic Gram-negative bacterium that can reach humans through direct contact with infected animals (lagomorphs, rodents, ticks, mosquitoes ...)
[Référence].
F. tularensis is classified as a Category A bioterrorism agent; indeed, some bacteria inhaled by aerosols are sufficient to induce severe acute pneumonia with a mortality rate of 30%. No effective vaccine is currently licensed and only a few broad-spectrum antibiotic compounds are used as drugs.
For many years, the Biology of Metal
team of the Chemistry and Biology of Metals laboratory has been studying the FUR*protein, a transcriptional metalloregulator whose activity is dependent on iron concentration and for which the team has developed inhibitors [1]. The multiplication and virulence of F. tularensis depend, among other things, on the importance of available iron stores in the host who, in order to defend himself, sequesters free iron. The response of the pathogen to this deprivation consists then in secreting a siderophore which makes it possible to mobilize the iron stored by the host. However, the expression of genes involved in the synthesis of this siderophore [Référence] is controlled by FUR.
A virulent strain isolated from a patient at Grenoble University Hospital (reference center of tularemia) suffering from a typhoid form of tularemia was used to characterize in vitro and in vivo the Fur protein of F. tularensis (FtFur). This work was done in collaboration with the TIMC-IMAG laboratory and the IBS. While the structure of FUR was to date reported as a dimer, researchers present the first structure of a tetrameric FUR protein consisting of two interlaced dimers [2. Moreover, this is the first time that a structure of FUR is presented as a pre-activated form containing iron, its physiological cofactor (Figure).
A coupled approach, developped by the Biology of Metal and BioCatalysis teams and the Laboratory's Modeling and Theoretical Chemistry group, used both modeling (free energy calculations) and experimental studies to provide evidence of a mechanism of tetrameric dissociation induced by specific DNA sequences. This phenomenon leads to the formation of a complex composed of two dimers attached to the DNA. The amino acid residues of FUR essential to this interaction have been identified.
The critical role of FtFur in pathogenesis, demonstrated in macrophages and in mice using a Δfur mutant with attenuated virulence, makes FtFur a promising anti-virulence target.
*
FUR (Ferric Uptake Regulator) is a bacterial protein that binds to DNA to control the expression of genes involved in iron homeostasis, virulence and oxidative stress.
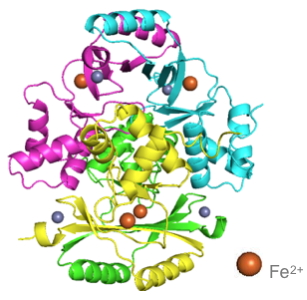
Three-dimensional structure of the FUR protein.
This work was made possible by A NRBC CEA financing, the GRAL and ARCANE Labex as well as the region and ARC Rhône-Alpes.