Unraveling, predicting and inhibiting protein-protein interactions involved in the maintenance of genome integrity.
Protein complexes lie at the heart of most biological processes. Our projects aim at unravelling how these physical interactions ensure proper cross-talks between cell machineries and signaling pathways enabling cells to resist to specific stresses. Currently, we focus on protein machineries involved in modulating the establishment of epigenetic information and acting in recombination processes. We combine experimental NMR and Xray crystallography methods to characterize the structure of protein complexes. We are also actively developing novel computational approaches of molecular docking to predict the structure of these complexes. Besides characterizing and predicting the structures of assemblies, we are also involved in the design of compounds that prevent the formation of complexes. In particular, we search for compounds inhibiting interactions triggered upon genotoxic stress to sensitize proliferating cells and improve the efficiency of anti-cancer treatments.
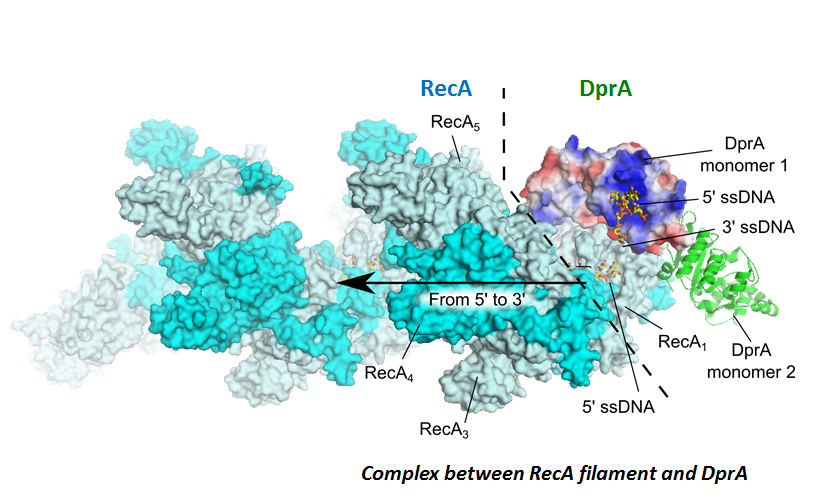
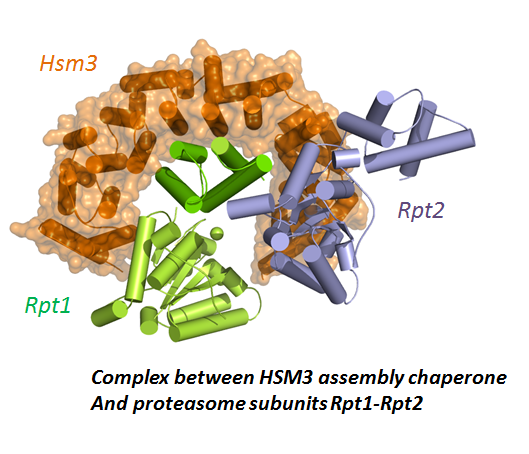
Structural characterization of complexes by integrative structural biology combining NMR, Xray crystallography and modeling.
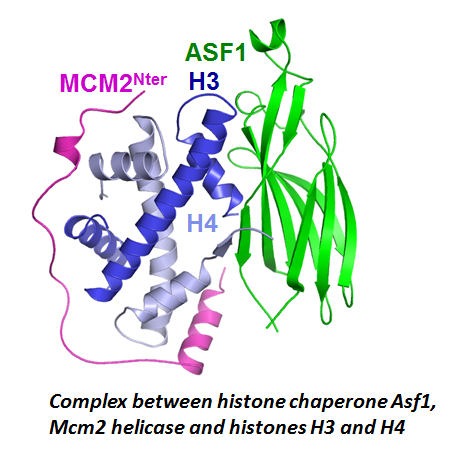 Maintenance of genome integrity relies on intricate cross-talks between DNA associated machineries, chromatin factors and signaling pathways regulated through versatile protein-protein interactions. Understanding the molecular logic underlying these interactions requires that competitive and synergistic associations be uncovered at different scales, from the atomic to the cellular one. We have been particularly interested in a class of proteins, called assembly chaperones which are involved in the formation of large macromolecular assemblies. As an example, we showed how Asf1, a central chaperone of histones H3 and H4, act in a synergistic manner with the Mcm2 helicase to coordinate the assembly and disassembly of nucleosomes at the replication fork. Other binding surfaces on Asf1 were also characterized revealing how other partners such as Rad53 (involved in signaling DNA damages), HirA (transcription) or CAF1 (replication) interact in a competitive manner with Asf1. These interactions take place in different epigenetic context and we are currently investigating how this context is influencing the network of interactions around Asf1.
Maintenance of genome integrity relies on intricate cross-talks between DNA associated machineries, chromatin factors and signaling pathways regulated through versatile protein-protein interactions. Understanding the molecular logic underlying these interactions requires that competitive and synergistic associations be uncovered at different scales, from the atomic to the cellular one. We have been particularly interested in a class of proteins, called assembly chaperones which are involved in the formation of large macromolecular assemblies. As an example, we showed how Asf1, a central chaperone of histones H3 and H4, act in a synergistic manner with the Mcm2 helicase to coordinate the assembly and disassembly of nucleosomes at the replication fork. Other binding surfaces on Asf1 were also characterized revealing how other partners such as Rad53 (involved in signaling DNA damages), HirA (transcription) or CAF1 (replication) interact in a competitive manner with Asf1. These interactions take place in different epigenetic context and we are currently investigating how this context is influencing the network of interactions around Asf1.
Predicting the structures of protein complexes using evolutionary information.

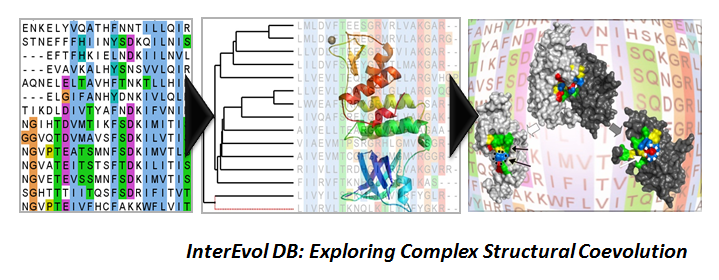
In the recent years, we have significantly contributed to improve methods for the prediction of complex structures by integrating coevolutionary information in the framework of the available docking tools. Through the development of the InterEvol database gathering a large number complexes of known structure, we first performed an in-depth analysis of the co-evolution process at their interfaces under a structural perspective. In particular we focused on more than a 1000 pairs of so-called interolog complexes (formed between homologous subunits) which allowed us to figure out important features at interfaces allowing for co-evolution. From this analysis we built the docking scoring function called InterEvScore by combining a two and three-body statistical potential with the coevolutionary information. These developments helped to achieve very good results for several targets proposed during the CAPRI experiment (Critical Assessment of Prediction of Interactions), in particular during the CASP11 conference in 2014. We are further trying to understand how we can improve the convergence and precision of our docking methods through a better exploitation of the wealth of the information contained in multiple sequence alignments.
Design of protein-protein interaction inhibitors
The inhibition of protein-protein interactions remains a significant methodological challenge which requires innovations in modeling, biomimetic chemistry, structural biology and cellular characterizations. Our team invested in this research field so as to inhibit the histone chaperone Asf1 and prevent its binding to its cognate partners the histones H3 and H4. Asf1 was recently recognized as a novel and promising anti-cancer target. We have developed a first generation of small peptides able to inhibit the interaction Asf1-histones. These peptides can penetrate cells efficiently, impede the cell cycle progression and trigger cell death in tumor cells cultures. Based on these encouraging results we are now tackling the conception of peptido-mimetic compounds, more powerful and selective, able to inhibit Asf1 activity in animals so as to demonstrate their therapeutic potential.