Project description
Eukaryotic cell membranes are comprised of several thousands of different lipid species. Why would cells generate such a repertoire if lipids were only fulfilling a barrier function? As a matter of fact, lipids are essential molecules, e.g. in cell signaling, as signposts for membrane identity, or for the regulation of membrane protein activity and membrane trafficking. Beyond diversity, lipids are characterized by a non random distribution over the two leaflets of cell membranes. This so-called transbilayer lipid distribution is regulated by three kind of lipid transporters (Figure 1), the flippases (transport toward the
inside), the floppases (transport toward the
outside) and the scramblases (transport in both directions).
The critical role of lipid asymmetry in cell physiology is illustrated by the fact that in humans, mutations of flippases (or P4-ATPases) result in severe neurological disorders, as well as a rare liver disease called progressive familial intrahepatic cholestasis. Furthermore, select flippases have been identified as strong contributors of virulence in the pathogen fungus Cryptococcus neoformans.
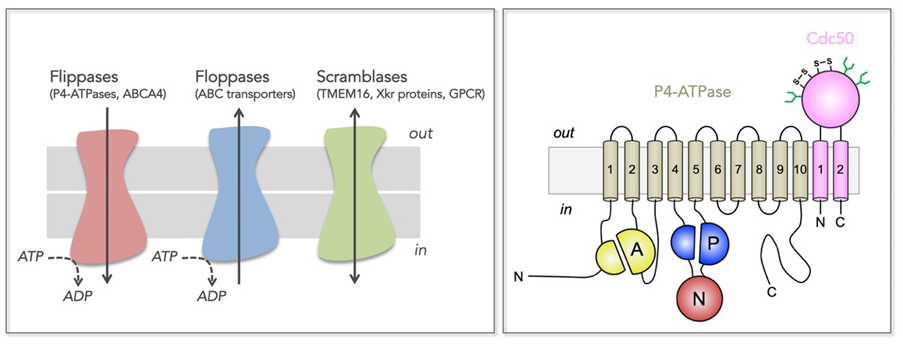
Figure 1: Transmembrane lipid transporters in eukaryotic cell membranes (adapted from Montigny et al, 2016).
Left, Flippases transport lipids from the exoplasmic (out) to the cytosolic (in) leaflet of membranes while floppases catalyze transport in the opposite direction. Scramblases disrupt phospholipid asymmetry by catalyzing a fast, bi-directional, energy-independent, and poorly specific transport. The arrows refer to the direction of lipid transport. Flippases and floppases use the energy of ATP to catalyze active transport.
Right, Topology of P4-ATPases and their associated CDC50 subunits, with the transmembrane domain in tan and the Actuator domain (A), the Nucleotide binding domain (N) and the Phosphorylation domain (P) in yellow, red, and blue, respectively. Cdc50 with two transmembrane spans and a large exoplasmic loop in pink; predicted disulfide bridges (S-S) and glycosylation sites (green) are indicated.
In this context, we aim to decipher the molecular mechanism of flippase-catalyzed transbilayer lipid transport, and the link between membrane remodeling and vesicle biogenesis. To this end, we devised a procedure for the co-expression and purification of the S. cerevisiae Drs2/Cdc50 flippase complex in a functional state (Jacquot et al, 2012; Azouaoui et al, 2014; Azouaoui et al, 2016) and identified phosphatidylserine (PS) and phosphatidylinositol-4-phosphate (PI4P) as key to the stability of the purified complex (Azouaoui et al, 2014). This allowed us to identify a critical role for the N- and C-terminal extensions of Drs2 in auto-inhibiting the complex and the contribution of PI4P in regulating its activity (Azouaoui et al, 2017). Using this pure and functional Drs2/Cdc50 complex, we recently obtained in collaboration with the laboratory of Poul Nissen (Aarhus University, Denmark) the first high-resolution structure of a lipid flippase (Timcenko et al, 2019) (Figure 2).
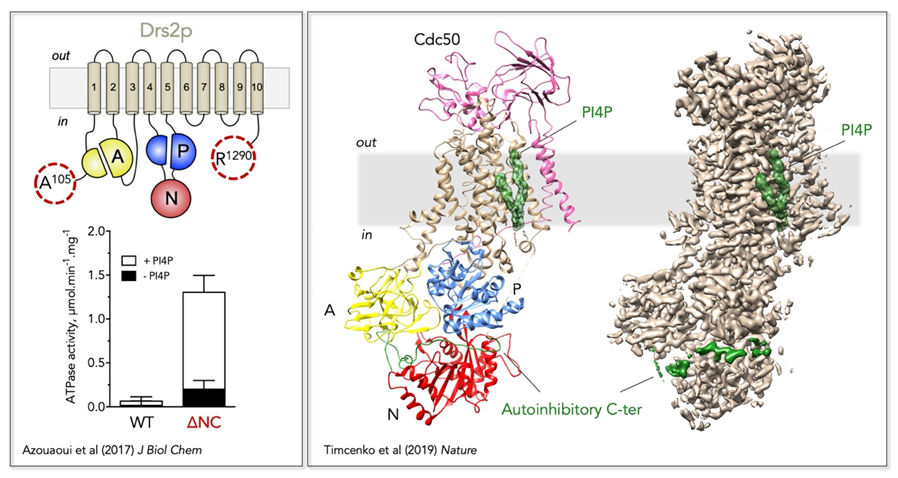
Figure 2: Regulatory mechanism of the yeast Drs2/Cdc50 phospholipid flippase.
Left, Trimming of the N- and C-terminal extensions of Drs2 dramatically increases its activity, which nevertheless remains dependent on PI4P.
Right,
Localization of Drs2/Cdc50 regulatory elements on its three-dimensional high-resolution structure (PDB: 6ROI, EMD: 4973). The Drs2 transmembrane domain is displayed in tan, the A domain in yellow, the N domain in red, and the P domain in blue on the structural model (left). PI4P is displayed as green surface and the autoinhibitory C-terminus as a green cartoon on the structural model. Both PI4P and the C-terminus are also shown on the EM map (right).
Both in the short and the long term, we develop the following projects :
-
Regulatory mechanism the Drs2/Cdc50 complex;
Although we know the complex is auto-inhibited by its terminal extensions, which part of the C-terminus is necessary for actual auto-inhibition remains to be elucidated. Also, what is the contribution of the N-ter in auto-regulation? Similarly, although the binding site for PI4P has been identified (Figure 2), the mechanism by which PI4P activates the flippase complex is unknown.
-
In vitro reconstitution of the lipid translocation machinery using purified components.
- Previous studies have shown that Drs2-interacting proteins Gea2 and Arl1 activate lipid transport in vivo. We want to characterize the interactions between Drs2/Cdc50, Gea2 and Arl1 in vitro as well as the possible requirement fo specific lipids.
-
Functional consequences of disease-causing mutations in ATP8B1.
- Mutations in the human P4-ATPase ATP8B1 have been linked to progressive familial intrahepatic cholestasis. We aim at establishing the purification of a functional ATP8B1/CDC50A complex and studying the consequences of ATP8B1 mutations on its activity.
-
Biochemical study of putative flippases from
C. neoformans.
- The P4-ATPase Apt1 is involved in C. neoformans virulence. First, is Apt1 a genuine lipid flippase? Then, we wish to screen small molecules in order to find inhibitors of the complex.
Recent publications
- Lyons JA, Timcenko M, Dieudonné T, Lenoir G, Nissen P (2020). P4-ATPases: how an old dog learnt new tricks – structure and mechanism of lipid flippases.
Curr Opin Struct Biol63:65
- Timcenko M, Lyons JA, Januliene D, Ulstrup JJ, Dieudonné T, Montigny C, Ash MR, Karlsen JL, Boesen T, Kühlbrandt W, Lenoir G*, Moeller A*, Nissen P* (2019). Structure and autoregulation of a P4-ATPase lipid flippase.
Nature571: 366.*senior co-authors
- Champeil P, de Foresta B, Picard M, Gauron C, Georgin D, le Maire M, Møller JV, Lenoir G, Montigny C (2019). Interaction of detergents with biological membranes: comparison of fluorescence assays with filtration protocols and implications for the rates of detergent association, dissociation and flip-flop.
PLoS One14: e0222932
- Lenoir G, Dieudonné T, Lamy A, Lejeune M, Vázquez-Ibar JL, Montigny C (2018). Screening of detergents for stabilization of functional membrane proteins.
Curr Protoc Protein Sci93:e59
- Azouaoui H, Montigny C, Dieudonné T, Champeil P, Jacquot A, Vázquez-Ibar JL, Le Maréchal P, Ulstrup J, Ash MR, Lyons JA, Nissen P, Lenoir G (2017). High phosphatidylinositol-4-phosphate (PI4P)-dependent ATPase activity for the Drs2p-Cdc50p flippase after removal of its N- and C-terminal extensions.
J Biol Chem 28792:7954
- Montigny C, Dieudonné T, Orlowski S, Vázquez-Ibar JL, Gauron C, Georgin D, Lund S, le Maire M, Møller JV, Champeil P, Lenoir G (2017). Slow phospholipid exchange between a detergent-solubilized membrane protein and lipid-detergent mixed micelles: brominated phospholipids as tools to follow its kinetics.
PLoS One12:e0170481
- Chaptal V, Delolme F, Kilburg A, Magnard S, Montigny C, Picard M, Prier C, Monticelli L, Bornert O, Agez M, Ravaud S, Orelle C, Wagner R, Jawhari A, Broutin I, Pebay-Peyroula E, Jault JM, Kaback HR, le Maire M, Falson P (2017). Quantification of Detergents Complexed with Membrane Proteins.
Sci Rep7:41751
-
Montigny C, Lyons J, Champeil P, Nissen P, Lenoir G (2016). On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim Biophys Acta 1861:767
-
Azouaoui H, Montigny C, Jacquot A, Barry R, Champeil P, Lenoir G (2016). Coordinated overexpression in yeast of a P4-ATPase and its associated Cdc50 subunit: the case of the Drs2p/Cdc50p lipid flippase complex. Methods Mol Biol1377:37