Iron-sulfur (Fe-S) enzymes have enormous and largely unexploited potential in biotechnology. They are essential for the biosynthesis of a wide variety of valuable natural products, such as biofuels, antifungals, antibacterials and anticancer drugs. Unfortunately, the dependence of Fe-S enzymes on their
cofactor often limits their catalytic activity. This problem creates bottlenecks in the biosynthetic pathways that depend on them. Even worse, most Fe-S enzymes exhibit little or no activity outside their native host, especially when expressed in a
heterologous organism. In the context of bioengineering, these obstacles are generally overcome. However, even if it is possible to express heterologous enzymes, it is difficult to guarantee the maintenance of their activity. Thus, the potential of Fe-S enzymes can only be fully exploited if we are able to routinely express them in active form in heterologous species.
Researchers at IRIG [
collaboration], have systematically studied the compatibility of prokaryotic Fe-S cluster assembly machinery (
Figure) of non-native Fe-S enzymes in
Escherichia coli, using bioinformatics, phylogeny, large-scale microbiology and biochemistry approaches. They focused on several Fe-S enzymes involved in metabolic pathways for the synthesis of valuable natural compounds (vitamin, biofuel, antibiotic). This systematic study identified several factors affecting the activity of heterologous Fe-S enzymes: oxygen sensitivity, incompatibility with the host Fe-S cluster biogenesis machinery, incompatibility with host electron transfer proteins, phylogenetic distance and likelihood to be functionally expressed, etc. Using collections of
orthologs Fe-S enzymes representative of the entire prokaryotic diversity, the researchers found a striking correlation between phylogenetic distance and the probability that an Fe-S protein will be functionally expressed. They also found that co-expression of a heterologous Fe-S biogenesis pathway increases the phylogenetic range of orthologs that can be supported by the foreign host. Finally, they also found that Fe-S enzymes that require specific electron transfer proteins to be active are rarely functionally expressed unless their specific reducing partners are identified and co-expressed.
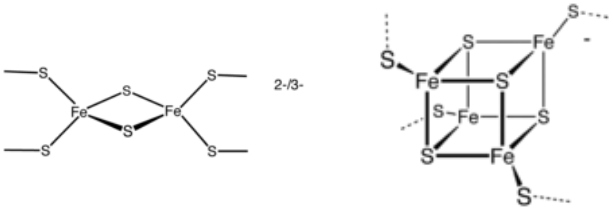
Two examples of Fe-S clusters. 2[Fe-2S] and [4Fe-4S] clusters.
The researchers applied these principles to improve the activity of an Fe-S enzyme from the bacterium
Streptomyces, involved in the biosynthetic pathway of an antibiotic, when expressed in
Escherichia coli. In particular, they identified a specific electron transfer protein that increases the activity of the Fe-S enzyme by a factor of 10, and the molecular basis of this specificity was identified.
This work as a whole clarifies how oxygen sensitivity and incompatibilities with foreign Fe-S center assembly and electron transfer machineries hinder the activities of heterologous Fe-S proteins. It shows how the identification of compatible electron transfer proteins and heterologous Fe-S biogenesis pathways is essential for the engineering of functional Fe-S enzyme-dependent metabolic pathways.
A cofactor is a non-protein chemical compound that is necessary for the biological activity of a protein. When this protein is an enzyme, the cofactor is involved in the catalytic reaction.
Iron-sulfur (Fe-S) clusters are inorganic cofactors composed of iron and sulfur atoms (Figure). They are present in proteins to which they confer numerous biological functions such as electron transfer, regulation of gene expression, catalysis, ... Fe-S clusters are built and distributed to Fe-S proteins and enzymes via complex multiprotein machineries. The essential components of these machineries are largely conserved and very versatile.
Heterologous organism: an organism used to produce a protein but which does not correspond to the original organism.
Ortholog. A gene (or protein) common to different species, originating from the same ancestral gene and having retained an identical structure and function during evolution.
Collaboration: Institut Pasteur Paris, Institute of Microbiology of Marseille and Delft University of Technology.