Baculoviruses are viruses with a circular double-stranded DNA (dsDNA) genome that specifically infect insect cells, playing an important role in regulating their populations. They are therefore widely used as biological agents in agriculture. In addition, they are used as expression systems, constituting a biotechnological tool of choice for the production of recombinant proteins in insect cell cultures.
Although commonly used, structural studies on the assembly of the
nucleocapsid* of baculoviruses at the molecular level remain limited. A better understanding of its organization would provide clearer insight into how it functions, especially regarding its effectiveness as an expression system.
In this study, researchers investigated the Baculovirus “Autographa californica multiple nucleopolyhedrovirus" (AcMNPV). This virus consists of a nucleocapsid surrounded by a lipid membrane in which viral glycoproteins are inserted. The nucleocapsid forms an elongated structure that is 50 nm wide and approximately 300 nm long on average, with two distinct terminal parts, the “apical" cap and the “basal" structure, which are connected to each other by the capsid. At the start of this study, many of the structural proteins that make up the nucleocapsid had not yet been localized or even identified.
Using cryo-ME, researchers focused particularly on the “basal" and “apical" structures of the nucleocapsid. They obtained several 3D maps ranging from high to medium resolution, allowing to identify and position proteins building each of these 3D maps. The AcMNPV nucleocapsid can be described by several distinct protein sub-assemblies, each with its own symmetry. By elucidating these different symmetries within these sub-assemblies for the first time, a pseudo-atomic model representing all the symmetrical parts of the AcMNPV nucleocapsid was obtained. To achieve this, at the highest resolutions of the 3D maps (below 4 Å), they were able to assign densities of unknown nature using “Modelangelo*". For 3D maps with lower resolution (above 4 Å), they identified and positioned several proteins based on structures predicted by “Alphafold*". AlphaFold was used not only to predict the structures of individual proteins among the 155 encoded by the AcMNPV genome, but also to suggest possible interaction partners, thus facilitating the interpretation of complex assemblies.
These results ultimately led to the identification of eight new proteins and a short segment of the viral genome in the “apical" domain of the nucleocapsid, providing experimental evidence for the proposed role of viral DNA entry and exit points.
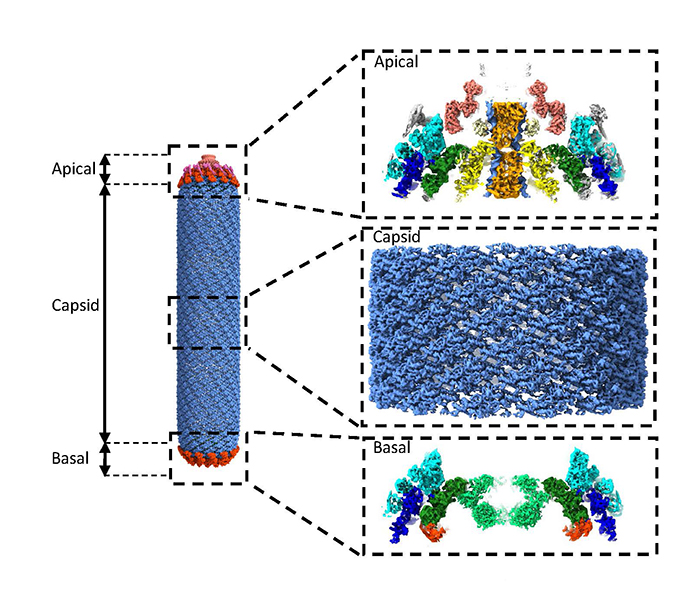
© CEA-Irig/IBS/G. Effantin
Structure of the whole nucleocapsid of AcMNPV obtained by cryo-EM. On the right, from top to bottom: zoom on a side view of the “apical” cap (the “entry point” for viral DNA), the capsid, and a side view of the “basal” structure.
Using high-resolution cryo-EM combined with AI algorithms, the authors of this study determined for the first time the entire structure—at near-atomic resolution—of the nucleocapsid of the Baculovirus AcMNPV. This work provides an in-depth understanding of how baculoviruses function by enriching the structural database of the virus, and will certainly contribute to a more rational development of biotechnological tools based on baculoviruses.
Nucleocapsid* : a complex consisting of the virus capsid and its nucleic acid (DNA or RNA), the viral genome.
Modelangelo* : machine learning program designed to build atomic models of proteins (with known or unknown amino acid sequences) from cryo-EM maps.
Alphafold* :
machine learning program that predicts the 3D structure of proteins based on their amino acid sequence. In 2024, Alphafold offered free access to more than 200 million protein structure predictions.
Collaborations :
European Synchrotron Radiation Facility (ESRF), Grenoble, France
European Molecular Biology Laboratory (EMBL), Grenoble, France