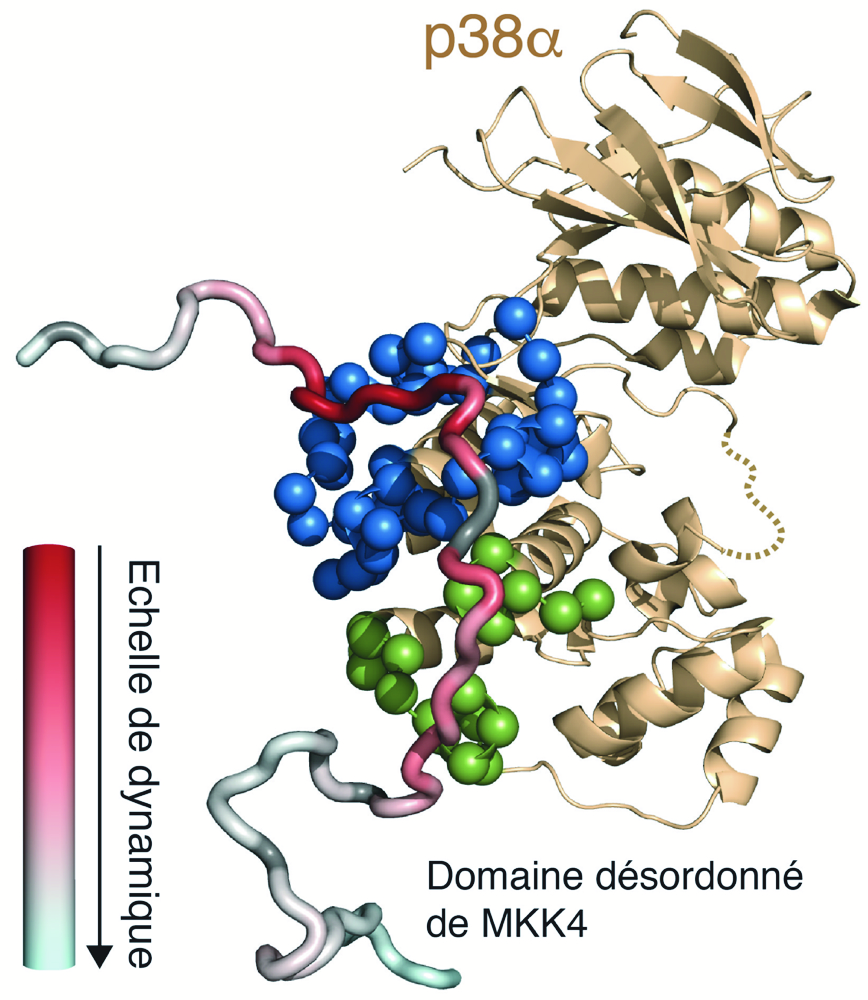
Intrinsically Disordered Proteins (IDP) are functionally active despite lacking a well-defined three-dimensional structure. Their great flexibility allows them to easily adapt to the surface of their partners and they are able to fold during interaction. In some cases, they may even form a so-called fuzzy complex, in which the IPD does not adopt a single conformation defined on the partner’s surface, but continues to sample multiple conformations in a highly dynamic complex.
The FDP group of the IBS, in collaboration with researchers from IAB Grenoble and the ENS Paris, succeeded in obtaining an atomic resolution description of the dynamics of an IDP in complex with its partner using nuclear magnetic resonance spectroscopy. Researchers applied this approach to the dynamic signaling complex formed between the mitogen-activated protein kinase (MAPK) p38α and the intrinsically disordered regulatory domain of the MAPK kinase MKK4.. The study shows that MKK4 uses a combination of interaction modes to link to p38α, leading to a complex displaying significantly different dynamics between linked regions.
The results show how IPDs can engage in very specific interactions without having a strong binding affinity.