Tumours are made up of
cells that proliferate excessively and
escape programmed cell death (apoptosis). One way of inducing the death of a tumour cell is
to break its DNA sufficiently. This is done in
radiotherapy and with certain chemotherapies. But tumour cells that have not been eliminated are capable of repairing the breaks in their DNA and can therefore proliferate again. A
complementary strategy is therefore
to act on DNA repair mechanisms.
Human cells repair their DNA in two different ways:
-
by joining the broken ends, known as the non-homologous end joining (NHEJ) system,
-
by copying the homologous sequence carried by a sister chromatid, known as homologous recombination.
The NHEJ pathway is the dominant repair system. After radiotherapy, tumour cells absolutely need this repair pathway, unlike normal cells. NHEJ is initiated by the Ku70/Ku80 protein complex, which rapidly encircles the ends of the break and acts as a tether for the other proteins needed to bind these ends together: DNA-PKc, Lig4, XRCC4 and XLF. The NHEJ system is highly regulated: cofactors, post-translational modifications, metabolites, etc
Around twenty years ago, scientists discovered that a small molecule produced by the body and sometimes used as a food supplement, inositol hexaphosphate (IP6) or phytic acid, binds to the Ku complex and stimulates NHEJ.
Using two techniques for studying proteins at the atomic scale (crystallography and cryo-electron microscopy), researchers from the I2BC (B3S department / INTGEN team, (UMR 9198, CNRS/CEA/UPSaclay, Gif-sur-Yvette), the IPBS (CNRS/Université Paul Sabatier) and the Institute for Structural and Chemical Biology (Leicester) have discovered how IP6 binds to the Ku protein. Biophysical experiments show that the binding of IP6 to Ku stabilises Ku's interaction with DNA and increases the affinity of the XLF protein for Ku. In vitro, in cell cultures, mutations in the IP6-Ku binding site reduce the accumulation of Ku at break sites and the recruitment of XLF to these same sites, thereby reducing the efficiency of the system. In other words, the presence of phytic acid on the Ku protein stimulates the attachment of the XLF protein to the Ku protein, an essential step in effectively sealing the DNA break (see Figure). This work opens up therapeutic prospects by identifying a new area on the Ku protein that could be targeted by small molecules to block break repair in tumour cells.
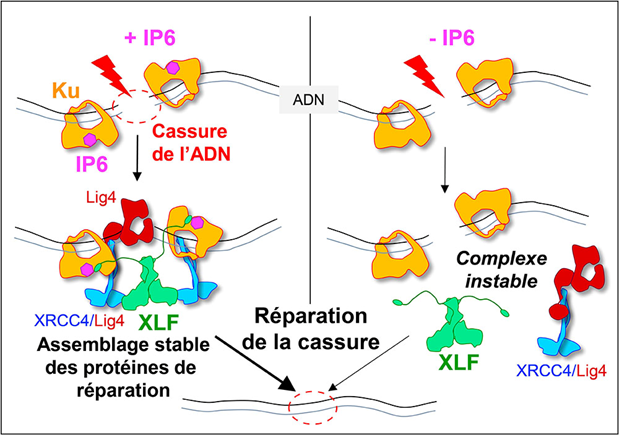
When a DNA double-strand break is induced, for example during radiotherapy, it is immediately recognised by the Ku protein, which serves as an anchoring site for the other proteins responsible for end-joining: DNA ligase 4 (Lig4), XRCC4 and XLF. The stability of this large protein complex depends on the binding of a small molecule, IP6, to Ku. When IP6 is absent (on the right of the model), XLF binds less well to Ku, the assembly is unstable and break repair is less efficient. © Ph. Frit / IPBS
Contact from Frédéric-Joliot institute for life sciences: