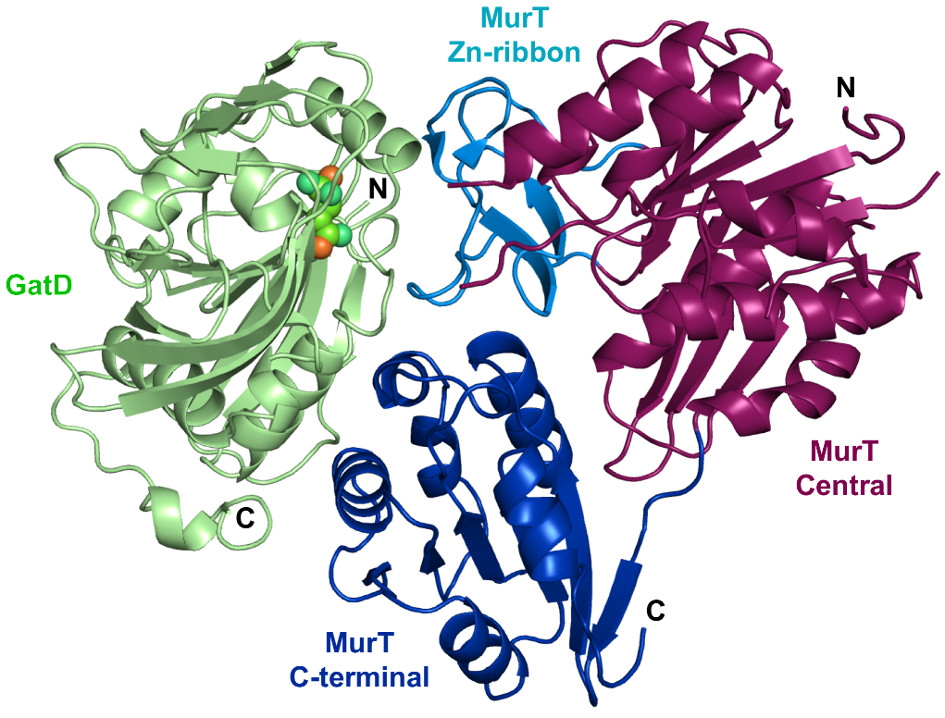
In some Gram-positive species such as Streptococcus pneumoniae, Staphylococcus aureus, or Mycobacterium tuberculosis, the second residue of the peptidoglycan precursor, D-glutamate, is amidated into iso-D-glutamine by the essential amidotransferase MurT/GatD complex. Here, IBS researchers in collaboration with the Norwegian University of Life Sciences, Newcastle University, Universidade Nova de Lisboa, Utrecht University and Université Paris Descartes, present the structure of this complex at 3.0 Å resolution.
MurT has central and C-terminal domains similar to Mur ligases with a cysteine-rich insertion, which probably binds zinc, contributing to the interface with GatD. The mechanism of amidation by MurT is likely similar to the condensation catalyzed by Mur ligases. GatD is a glutaminase providing ammonia that is likely channeled to the MurT active site through a cavity network.
The structure and assay presented here constitute a knowledge base for future drug development studies.