A bacterium is Gram-positive or Gram-negative following its reaction to a staining test called the Gram test. This is the first test to be performed when trying to identify the bacterium responsible for an infection during a bacterial diagnosis.
In collaboration with EPFL (Switzerland), we have developed an optical trapping device based on the use of a 2D hollow photonic crystal cavity dedicated to the study of bacteria. The trapping of seven types of bacteria has been demonstrated on these optical structure:
Pseudomonas putida,
Neisseria sicca,
Escherichia coli,
Yersinia ruckeri,
Bacillus subtilis,
Listeria innocua and
Staphylococcus epidermidis.
The trapping signal of these bacteria transmitted through the optical structure is characterized by two phenomena: a sudden variation of the transmitted intensity and strong oscillations related to the displacement of the bacterium when it interacts with the cavity field. We show that the combination of these two characteristics allows the identification of the Gram type of the studied bacteria and sometimes to their absolute identification among this sample of seven bacterial species.
This technique therefore makes it possible to identify the Gram type of bacteria by a fast label-free method, working at a low concentration of bacteria. Moreover, this method is non-destructive, unlike the classical Gram test, it is therefore possible to use the same cells to perform further analyses, promoting their identification.
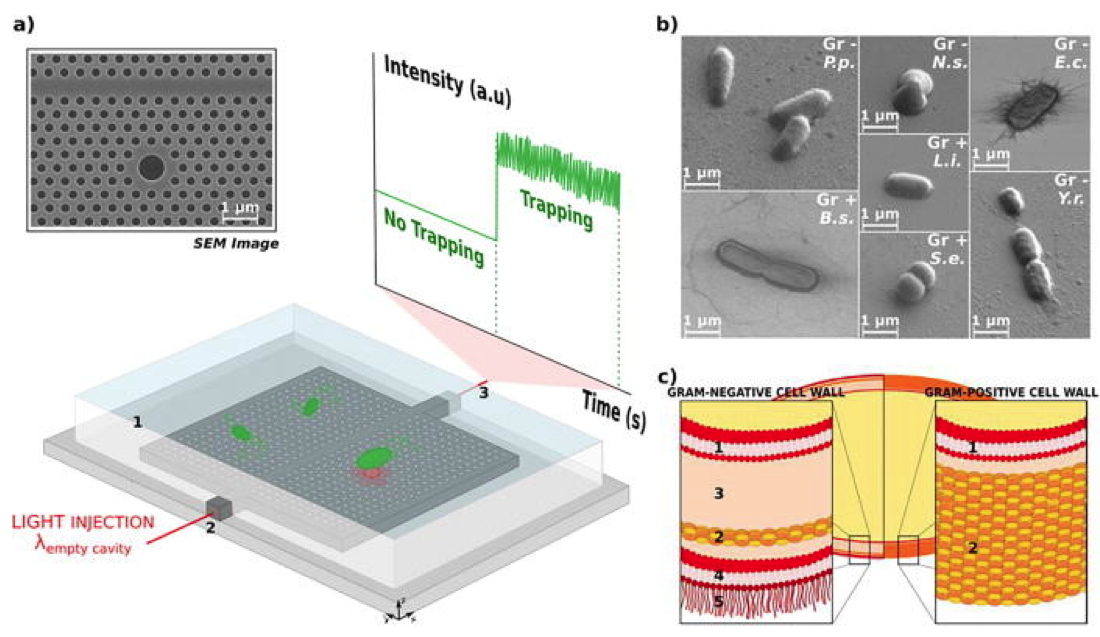
Figure : (a) SEM image of the 2D hollow photonic crystal cavity and experimental setup. The photonic crystal structures are immersed in water (1) thanks to a PDMS frame that allows for the Brownian motion of bacteria in the proximity of the optical cavity. Light from a tunable laser is injected at the resonance wavelength into the access waveguide (2) in an end-fire setup. The transmitted light is collected from the other facet of the sample (3)
via a microscope objective and its intensity is monitored with an oscilloscope. (b) SEM pictures of the seven bacteria under study. P.p. stands for
Pseudomonas putida, N.s. for
Neisseria sicca, E.c. for
Escherichia coli, Y.r. for
Yersinia ruckeri, B.s. for
Bacillus subtilis, L.i. for
Listeria innocua, and S.e. for
Staphylococcus epidermidis. The Gram-type of these bacteria is also indicated: Gr - for Gram-negative and Gr + for Gram-positive bacteria. (c) Depiction of the structural differences in the cell wall for Gram-negative and Gram-positive bacteria. Gram - bacteria show two membranes (plasma 1 and outer 4) separated by a liquid periplasmic space (3) and by a thin (5–10 nm) peptidoglycan layer (2). Moreover, lipopolysaccharides (LPSs, 5) project from the outer membrane. On the contrary, Gram + bacteria exhibit a less complex structure, with a single membrane (plasma 1), surrounded by a thick cell envelope (20–80 nm) made of a peptidoglycan layer (2) and no LPSs.