Super-resolution fluorescence microscopy makes it possible to observe living matter at the nanoscopic scale from both structural and dynamic point of views. In the latter case, individual target molecules are tracked when they diffuse into a cell, using a technique called
sptPALM (single-particle-tracking Photo-Activated Localization Microscopy). However, a major obstacle to this technique is the imperfection of the fluorescent proteins used as markers, which tend to "blink", i.e. to fade transiently, easily causing the loss of the individual molecule's tracks. Is there a strategy to reduce or eliminate this problem?
Researchers at our institute, in collaboration with the Catholic University of Leuven in Belgium, have undertaken to study the nature of blinking in mEos4b, the latest variant of a series of green to red photoconvertible fluorescent proteins widely used in super-resolution microscopy.
Using a mechanistic approach combining X-ray crystallography, optical spectroscopy and analysis of the fluorescence traces of single molecules, the work revealed a major source of blinking, linked to a photochromism phenomenon. Following excitation by the 561 nm laser of the sptPALM microscope, fluorescent proteins can change conformation and transiently convert into a non-fluorescent state. The study of this non-fluorescent state then revealed its high sensitivity to cyan-coloured light. Thus, a low illumination of the sample with a cyan laser at 488 nm forces a rapid return to the fluorescent state, considerably reducing the lifetime of the non-fluorescent state, and consequently the intensity of blinking.
Since most sptPALM instruments have a 488 nm laser, additional illumination at this wavelength is very easy to achieve, providing a significant improvement in data quality with minimal effort. In their paper, the researchers were able to study the diffusion of the MAP4 protein (microtubule associated protein 4), which interacts with microtubules, in a much more precise way than before. They are now trying to apply their flicker suppression strategy to the issue of molecular counting by qPALM (quantitative Photo-Activated Localization Microscopy) imaging.
In this work, the mechanistic understanding of the origin of the phenomenon of mEos4b blinking, using advanced structural biology techniques, has provided a simple solution to a microscopy problem in cell biology. This is therefore a case study demonstrating the full methodological potential of an integrative approach between structural and cell biology.
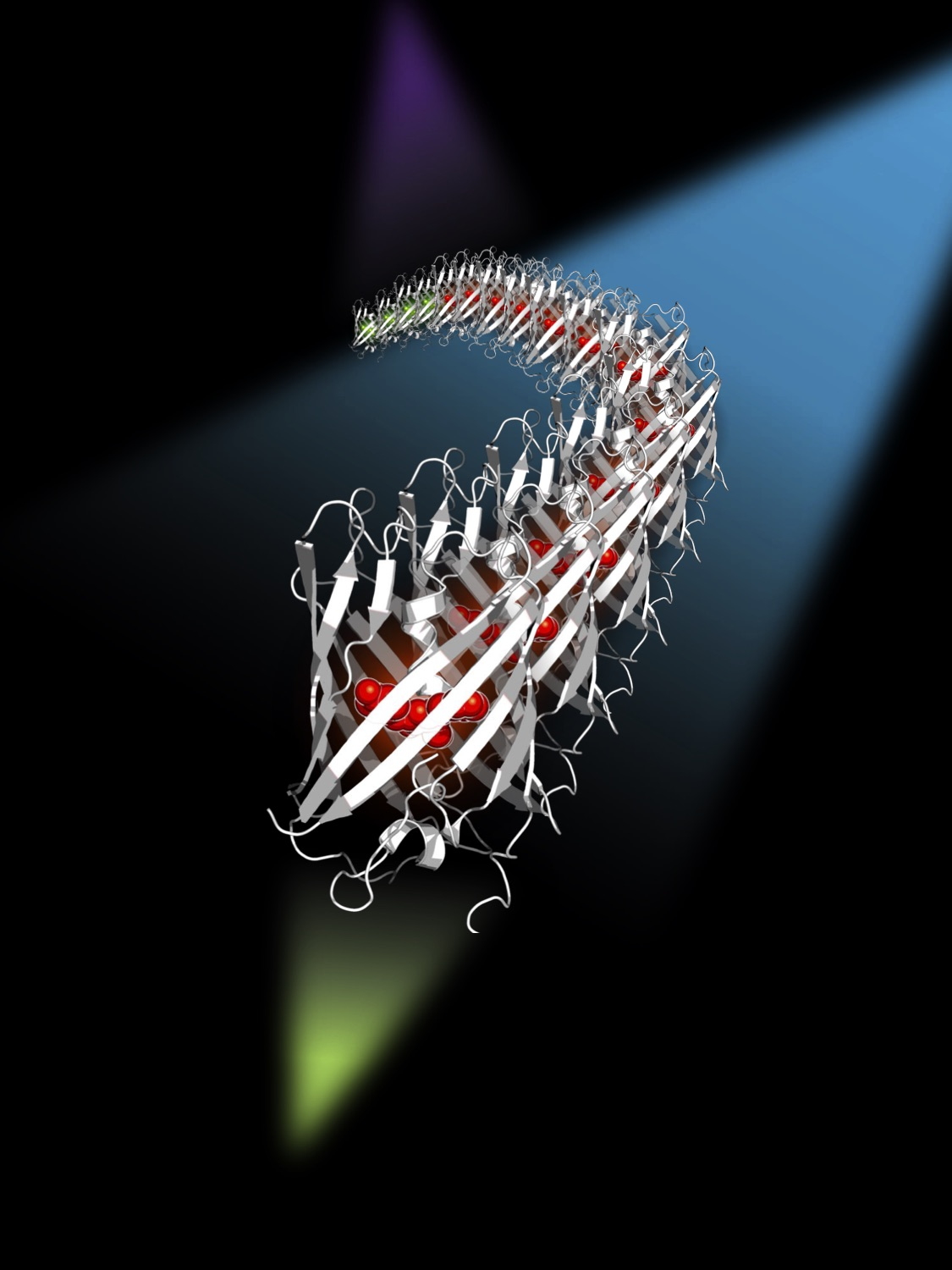
Artist's view of the mEos4b protein diffusing in a sample, and illuminated by three lasers at 405 nm, 488 nm and 561 nm. The 488 nm laser suppresses the blinking of mEos4b.
sptPALM: in this technique, the target molecules are most often labelled with a “green-to-red photoconvertible” fluorescent protein. The individual fluorescent proteins, initially in the green state, are gradually photoconverted to the red state using a 405 nm laser, and the red fluorescence emission is visualized over time using a high sensitivity wide field microscope equipped with a 561 nm excitation laser, allowing the trace of each target molecule to be followed.