The development of any multicellular organism (plant or animal) depends on genes that must be activated in the right tissue at the right time. The activation of these genes is controlled by proteins called
transcription factors*. Auxin Response Factors (ARFs) are among these factors: by activating or inhibiting a multitude of genes, they enable auxin, a plant hormone, to play roles that are always important but differ depending on the plant tissue (for example, guiding root growth according to gravity, causing a flower to emerge, or helping the stem to grow toward the light).
How does the same “auxin" signal enable this multiplicity of responses: activation and inhibition of genes specific to each tissue ?
Together with their collaborators, researchers at
CEA-Irig/LPCV-Flo_Re have attempted to understand how auxin and ARFs control gene activity in Arabidopsis in space and time.
In Arabidopsis, there are 23 ARF proteins, some considered activators and others inhibitors. Until now, it was assumed that the response to auxin depended on the ARFs present in a given tissue, which competed to activate or repress each gene.
But the solution is more complex. ARFs act by binding to DNA motifs located near the genes they control. By combining biochemical approaches, single-cell sequencing, and "in planta" studies, the study reveals that the diversity of responses to auxin also depends heavily on the combinations of motifs present near each gene.
Using synthetic sequences consisting of different motif configurations, the researchers discovered that each ARF has a preference for certain configurations, and that its activity—as an activator or repressor—varies depending on the arrangement of the motifs. The action of auxin therefore involves two levels: on the one hand, the combination of ARFs present in each cell and, on the other hand, the configuration of motifs present in each gene. This combination generates a complex “two-layer" ARF/motif regulatory code.
This two-level code allows each gene to respond “in its own way" and according to the tissue, and plays a major role in the diversification of responses to auxin in plant tissue development.
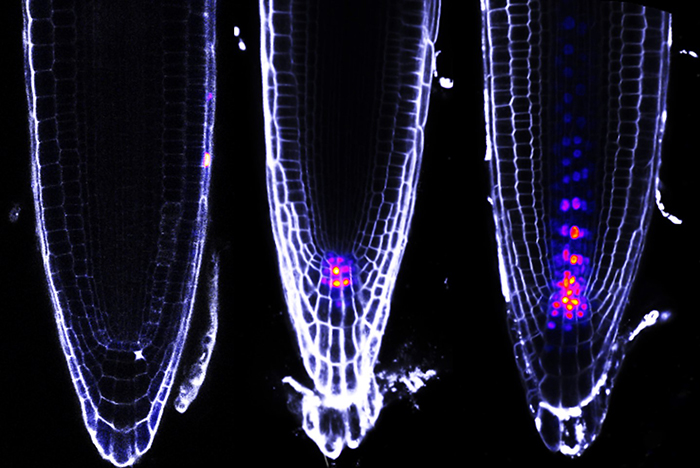
© CEA-Irig/LPCV/Flo_RE/R. Dumas
Activity in Arabidopsis roots of three synthetic DNA sequences constructed from three different configurations bound by ARFs and controlling the expression of a fluorescent protein (mTurquoise). This figure illustrates that varied expression profiles give rise to a double-layer code based on the composition of ARFs in each cell and on the binding of ARFs to different DNA motif configurations. The images were obtained by confocal microscopy. Fluorescence is visualized in a color range from violet to yellow-orange. Images from Raquel Martin-Arevalillo.
Understanding how, within an organism, information from a developmental signal is translated into a multitude of cellular responses in space and time is a key factor in understanding the development of that organism. Thanks to this study, it is now possible to understand how auxin can induce a multitude of transcriptional responses, despite the complexity of the molecular interactions involved. This breakthrough opens up new prospects for agriculture and medicine, by enabling better control and prediction of organisms' responses to developmental signals.
Transcription factors* : proteins capable of binding to DNA and regulating genes by activating or inhibiting mRNA synthesis.
Fundings : ANR ChromAuxi 18-CE12-0014-02, ERC TEMPO Project 101095380, GRAL/CBH-EUR-GS (ANR-17-EURE-0003)
Collaborations :
Laboratoire Reproduction et Développement des Plantes, Université de Lyon, ENS de Lyon, CNRS, INRAE, INRIA, 69342 Lyon, France
Institute of Synthetic Biology, University of Düsseldorf, 40225 Düsseldorf, Germany
CEPLAS – Cluster of Excellence on Plant Sciences, University of Düsseldorf, 40225 Düsseldorf, Germany
Center for Genomics and Systems Biology, New York University, New York, NY, USA
Center for Genomics and Systems Biology, New York University Abu Dhabi, Abu Dhabi, United Arab Emirates