To meet the needs of clinical diagnosis and microbiological screening in general, it is essential to be able to quickly and reliably identify the microorganisms present in a microbiota sample (normal or pathological microbial flora). In order to better understand the individual components of microbiota, culturomics is being developed as a means of generating numerous microbial isolates with highly distinct characteristics by systematically testing hundreds of different culture conditions. High-throughput identification methods now need to be developed to rapidly screen these isolates.
In the present proof of concept, the researchers propose an innovative multiplex proteotyping methodology for the simultaneous analysis of 21 bacterial samples by tandem mass spectrometry, without relying on differential peptide labeling that would require costly chemical reagents. The method is based on pre-fractionation of the peptidomes of each microbial isolate by reverse-phase chromatography, mixing of specific fractions of each isolate, differentiated according to their hydrophobicity, followed by analysis of the mixture in a single nanoLC-MS/MS (Liquid Chromatography coupled to tandem Mass Spectrometry) experiment. Organism-specific signals are distinguished from other signals by their hydrophobic properties, and the process identified all 21 microorganisms in a single 60-minute analysis. The researchers also developed a specific proteotyping concept to assign taxonomic information to signals restricted by time unit, thanks to clever phylopeptidomic computer processing.
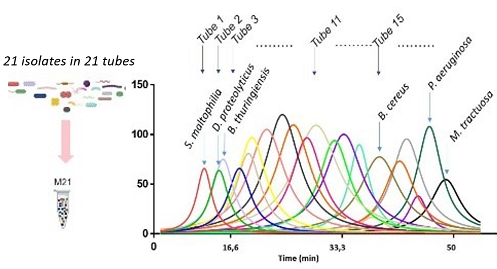
Schematic diagram of the experimental protocol and analysis results © J
Armengaud / CEA
Conclusion: By developing a label-free multiplexing strategy for the analysis of microbial isolates by tandem mass spectrometry, and in view of recent technological advances in this approach, the authors of this study envisage being able to identify up to 200 microbial isolates per hour, enough to revolutionize "culturomics" and advance our knowledge of microbiota.
Contacts : Béatrice Alpha-Bazin (beatrice.alpha-bazin@cea.fr) – Jean Armengaud (jean.armengaud@cea.fr)
- Proteotyping is based on the analysis and identification of peptides by mass spectrometry.
- The microbiota is the set of microorganisms - bacteria, viruses, parasites and non-pathogenic fungi, known as commensals - that live in a specific environment. In humans, the intestinal microbiota is the most "populated" of these, harboring 1012 to 1014 microorganisms.
- Phylopeptidomics is a mathematical method developed at Li2D to identify the contribution of each of the organisms that make up a microbiota in a metaproteomics dataset.
- Culturomics involves the multiplication of culture media and conditions, and the rapid identification of bacteria by mass spectrometry. In particular, it has enabled the discovery of species never isolated previously in humans, as well as new bacterial species or genera. In this respect, it is highly complementary to metagenomics, which does a relatively good job of assessing the presence of different majority flora, particularly bacteria that are not yet cultivable, whereas culturomics is better at assessing the diversity of minority flora, dominated by a few over-represented species.