| Genome integrity is constantly threatened by DNA lesions. Unrepaired or incorrectly repaired DNA damage may lead to loss of heterozygosity, mutations, deletions, genomic rearrangements and chromosome loss. Such genetic alterations are the main causes of cancer and other genetic diseases. Consequently, DNA repair processes have to be tightly regulated. The three dimensional organization of the genome in the nucleus, previously known to impact on gene expression, has recently emerged as a key regulator of DNA repair.
Double strand breaks signaling and repair. DSBs are first detected and signaled by the DNA damage checkpoint that triggers cell cycle arrest, providing time for the cell to repair damaged chromosomes before entering mitosis. Unless a DSB is faithfully religated by Non-Homologous End Joining (NHEJ), it is processed into 3’ single strand overhangs through the action of partially redundant nucleases. Resection is accompanied by the binding of replication protein A (RPA) to the 3’ single-stranded overhangs, which helps recruiting the checkpoint complexes. Once recruited to DSB, these complexes get activated and induce the phosphorylation of numerous targets including transducing kinases, which subsequently phosphorylate downstream effectors to delay cell cycle and promote DNA repair. Additionally, the checkpoint kinases modify the chromatin surrounding DNA damages through phosphorylation of the H2A histone (H2AX in mammals). Although phosphorylation of H2A represents a major histone modification that functions in DNA repair, a growing body of literature has implicated additional histone modifications. In all organisms, two different mechanisms are used to repair DSBs: Homologous recombination (HR) and non-homologous end joining (NHEJ). NHEJ simply religates the broken ends whereas during HR a DNA break is repaired by copying homologous sequences present elsewhere in the genome. HR comprises different pathways: gene conversion (GC) that accurately repair the lesion by copying the homologous sequence, single strand annealing (SSA) or break induced replication (BIR) that can both lead to loss of genetic information. In general, NHEJ is the preferred pathway in the G1 phase of the cell cycle, while HR is favored in S and G2 phases. Indeed in S and G2 phase, RPA binding to 3’ single strand overhangs facilitates the recruitment of proteins of the Rad52 epistasis group, among which Rad51, which carries out the strand-exchange reaction. Whereas the different DNA repair pathways and the proteins they involve are know pretty well described, how they are regulated relative to each other in time and space remains to be deciphered. It seems now clear that posttranslational modification of both DNA repair and checkpoint proteins is of importance for the regulation of their activities but how these modifications are regulated and how they affect the activity of the proteins only begins to be described.
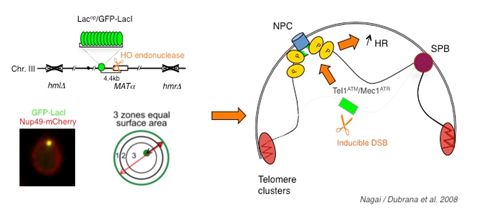 Double strand breaks localize to the nuclear periphery. Double strand breaks localize to the nuclear periphery. A haploid strain lacking HM loci and containing a galactose inducible HO endonuclease was used such that a single permanent double strand break is created on galactose containing medium. The MAT locus was tagged by insertion of 256 lac operators at 4.4 kb of the HO cut site. The lacop array is visualized by binding of a GFP-lac-repressor fusion and the nuclear envelope is visualized through a Nup49-mCherry fusion. Change in localisation depends on the checkpoint kinases Tel1ATM and Mec1ATR and has a positive effect on spontaneous recombination.
Nuclear organization During the last decade, the non-random spatial arrangement of the genome into the nucleus of eukaryotic cells, as emerged as a key regulator of genome functions and notably of the propagation of a stable genome. One feature of nuclear organization is the existence of subcompartments in which specific DNA sequences and proteins associate creating microenvironments that can be more or less favorable for specific processes. Concerning the maintenance of genome integrity, recent data highlight the importance of nuclear architecture. In mammals, mutation of proteins involved in the nuclear architecture (lamins and lamin associated proteins) results in diseases associated with genomic instability. A strong correlation between spatial proximity of chromosomes/genes in interphase nuclei and translocation frequencies has been observed. During the last few years, studies in yeast further revealed the importance of nuclear organization for the maintenance of a stable genome. Notably, we and others recently showed that an irreparable or slowly repaired DSB localizes to the nuclear periphery. Several lines of evidence implicate nuclear pore complexes and the inner nuclear membrane Mps3 as binding sites for the irreparable DSB. We further showed by Chromatin Immunoprecipitation (ChIP) that an irreparable DSB becomes associated with the Nup84 pore subcomplex and with the Slx5/Slx8 heterodimer, both complexes being colocalized at the nuclear periphery. The Slx5 and Slx8 proteins form a conserved SUMO-dependent ubiquitin ligase complex that plays an important role in the maintenance of genome integrity possibly by targeting sumoylated proteins to the proteasome. In addition, the Slx5/Slx8 complex has been shown to counteract Rad51-independent recombination, such as single-strand annealing and the break induced replication. Altogether these data suggest a model in which targeting repair intermediate to the nuclear pore may influences the choice of the repair pathway through steps that are controlled by SUMO recognition, ubiquitylation, and degradation. |