The HARDCORE of the double-strand break repair machinery
Monitoring the health of the DNA is the job of the many repair systems present in our cells, each specialised in detecting and correcting a specific anomaly. A particularly serious anomaly is the
breakage of the double-strand DNA, which occurs as a result of oxidative stress or exposure to radioactivity, X-rays (X-ray examination) or cosmic radiation (during air travel). In human cells,
the maintenance service for this type of break is called "NHEJ" (Non Homologous End Joining), which coordinates numerous agents to detect breaks, "isolate" them and finally join them back together again.
The hard core of this process is made up of Ku70/Ku80, DNA-PKcs, Lig4, XRCC4 and XLF. Thanks to its buoyancy, the Ku70/Ku80 complex quickly encircles each end of the break and acts as a mooring for a DNA-PKcs enzyme. The two DNA-PKcs, each at one end of the break, then join forces.
In a previous study, teams from
B3S (I2BC/Institut Joliot) and the Institute of Pharmacology and Structural Biology - IPBS - (in collaboration with Gustave Roussy and the Universities of Paris-Saclay and Aix-Marseille) described at the atomic level how:
PAXX, not that redundant
Of all the other 'accessory' players involved, the
PAXX protein is the most recent, having been identified in 2015. PAXX
also interacts with Ku70/Ku80 (on Ku70) and its function appears to be partially redundant with that of XLF. Using two techniques that allow proteins to be visualised at the atomic level (X-ray crystallography and cryo-electron microscopy), the B3S and IPBS researchers precisely identified the contact zones between PAXX and Ku70/Ku80 and showed that
PAXX and XLF can simultaneously bind to Ku on opposite sides (PAXX to Ku70 and XLF to Ku80, Figure 1).
PAXX and XLF act as bridges, stabilising alternative forms of DNA-PKcs dimers. The assembly of these different macromolecular assemblies may provide
a means for the cell to repair DNA according to the nature of the DNA breaks. . As mentioned above, PAXX is an accessory protein in the NHEJ process and its loss only moderately affects the process. In this study, the researchers show that deletion of PAXX exacerbates the effect of XLF loss.
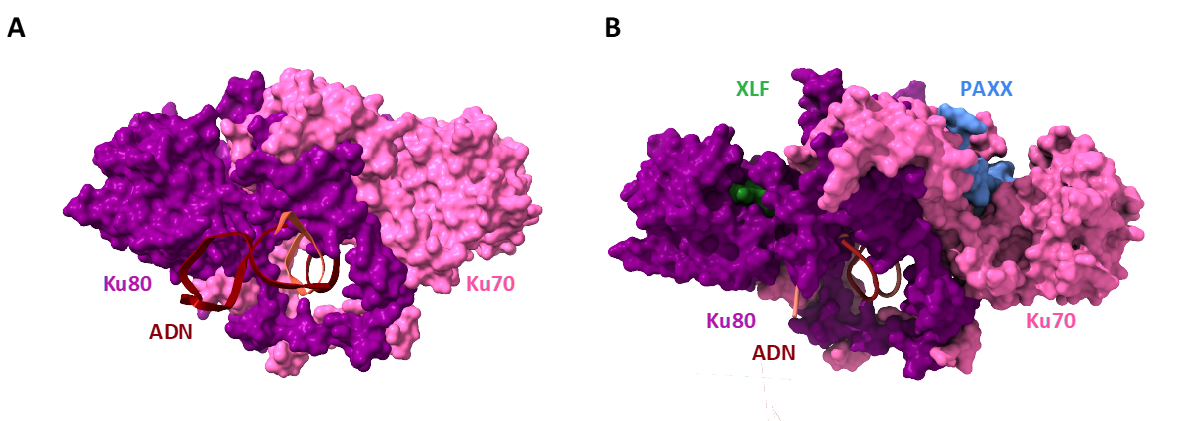
Figure 1 : The Ku heterodimer, a dynamic ring that spreads its wings like a butterfly to attract its partners.
A. Atomic structure of Ku bound to DNA, determined by X-ray crystallography. DNA, in brown, is bound to the Ku ring composed of the two subunits Ku80, in magenta, and Ku70, in pink.
B. Structure of Ku bound to two repair factors: XLF, in green, binds to the Ku80 subunit. PAXX, in blue, binds to the Ku70 subunit. Ku unfolds on both sides like a butterfly when the two factors bind. © Seif-El-Dahan / I2BC
Hyperactive cancer cells are particularly susceptible to DNA breaks induced by radiotherapy;
molecules that block the assembly of PAXX and/or XLF at DNA breaks could increase the sensitivity of cancer cells and thus open up new therapeutic prospects. Initial work coordinated by B3S has identified some promising leads.
Contact Institut Frédéric-Joliot :
[1] Cette fois-ci, en collaboration avec le département de biochimie de l'Université de Cambridge, AstraZeneca et le Leicester Institute for Structural and Chemical Biology