Brain imaging genomics, recently emerging field of research, tries to relate brain phenotypes to the even richer genomic-wide molecular data available through sequencing: genotype, methylation and CNV from blood and even transcriptomics and proteomics in some studies.
Since years 2010, we have been involved in two major European imaging-genetics cohorts, namely IMAGEN and AIMS-TRIALS. We participated in several collaborations devoted to study associations and potential causalities between genomic variants, endo-phenotype (imaging features) and developmental/psychiatric phenotypes. Leveraging our long-term experience in imaging-genetics, we started several projects in the UK BioBank cohorts. We studied the original sulcal phenotypes developed at NeuroSpin for years. Starting from functional imaging for the “Language Networks”, we found associations in several genomic regions that may drive specific aspects of neuro-development in human fetus. Conversely, starting from “Human Gained Enhancers” loci of the genome, we found associated regions that may
enlighten the effects in the brain of the evolution in our hominin ancestors (see Lemaitre et al.). In future work, we will address the unveiling of the multi-variate implication of genomics and environment in the normal and pathological development of the brain in both early and aged life through multi-omic multi-modal integration. This integration requires novel frameworks that are emerging in machine or deep learning like RGCCA or multi- channel VAE.
Based on our recent works on multi-block integration in radio-genomics (oncology) or in imaging-genetics (autism), we will continue to develop models able to take into account the conditions of data collection in population studies, in particular the stratified organization of patient groups. Another major challenge of the developments in integration will concern the causality. We plan to apply these developments in Alzheimer's Disease, Autism or Depression by characterizing several development trajectories in ageing and early development (cohorts HCP, UK BioBank, ABCD, Memento).
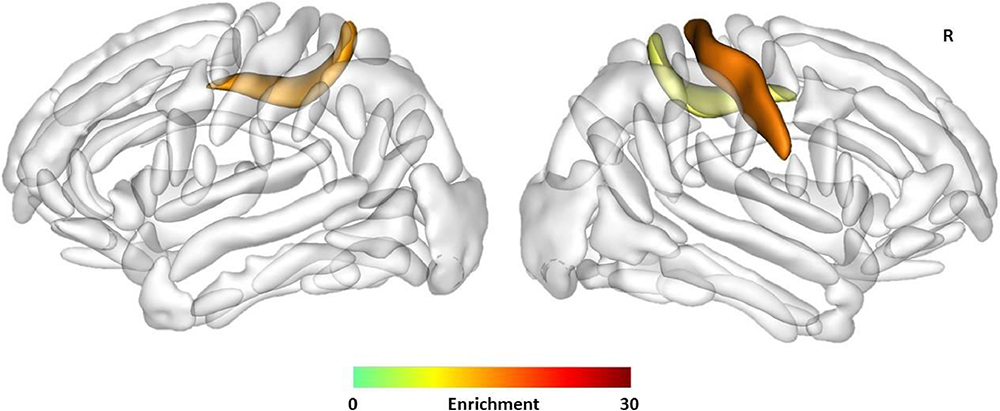
Enrichment of heritability of sulcal opening in the left (Enrichment = 16.54, FDR corrected P-value = 0.040) and right calloso-marginal posterior fissures (Enrichment = 22.44, FDR corrected P-value = 0.040) and in the right central sulcus (Enrichment = 19.33, FDR corrected P-value = 0.034) with the Human Gained Enhancers (HGEs) active at 7 weeks postconception. The sulci are displayed using the statistical probability anatomy map (SPAM) representation, which represents the average sulci shape and position on the reference base of the BrainVISA, R: Right. ©Neuroimage, Elsevier