After an extensive inventory of species described in the literature, we noticed that the species Acinetobacter baylyi, which was isolated from soil, presents real advantages as a model for the study of bacterial metabolism. A. baylyi has the highest capacity for transformation by exogenous DNA (natural competence (1)).
of any micro-organism. This property, associated with its capacity to undergo homologous recombination (2), makes genetic modification very easy. This is a process of targeted exchange (directed mutagenesis) which permits specific modification of genetic information at a given locus.
A. baylyibelongs to the gamma proteobacteria like E. coli with which it shares 46% of its genes. Furthermore, it has several decisive advantages compared with E. coli : natural competence, faster growth on minimal media and no metabolic ambiguity (strictly aerobic).
Development of resources for the systematic study of the A. baylyi GENOME
Sequencing and annotation of the genome
With the goal of developing a genetic model which can be manipulated more easily
thanE. coli, we have undertaken the systematic analysis of the genome of A. baylyi in order to gain a more complete and detailed knowledge of its metabolic functions. With this objective, the sequencing of the genome has been performed at Genoscope, followed by annotation with the participation of experts on bacterial metabolism (V. Barbe & al., 2004). Of the 3205 CDS (
3) annotated, 36% have a known function, 28% a "probable" function and 36% have no known function. Furthermore, this first analysis demonstrated that the
A. baylyigenome, which is more simple than that of
E. coli, includes fewer gene duplications. This should simplify the phenotypic analysis of mutants considerably. A special effort was made in the reconstruction of the metabolic pathways of this bacterium.
Establishment of a collection of replacement mutants
The systematic analysis was followed by the establishment of
a collection of replacement mutants for each CDS which was identified in the course of annotation of the genome. Of 3,205 CDS, 2,594 replacement mutants were obtained (81%). We were not able to obtain mutants with a correct structure for 499 genes which are therefore candidate essential genes for ADP1 life on minimal medium
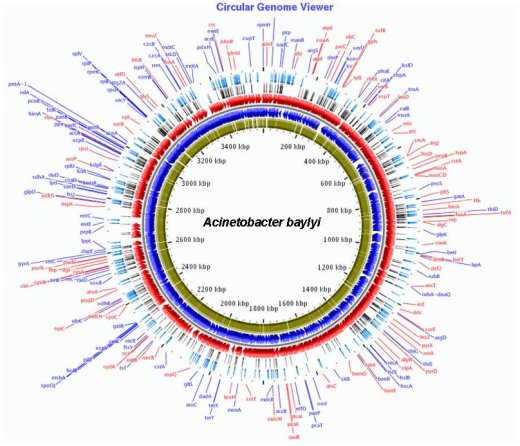
Circular representation of the genome of A. baylyi (Red and blue): genes localized on the (+) and (-) strands. (Green): genes for which a mutant has been obtained. (Black): Genes for which no mutant was obtained. (Light blue): Enzymes of a biosynthetic pathway.
Glossaire :
(
1) Competence (of a bacterium) : Capacity of a bacterium to incorporate foreign DNA.
(
2) Homologous recombination : Process of exchange between two identical regions of DNA.
(
3) CDS : Coding DNA Sequence.
Mutant collection
Principle and Method :
Each CDS (
1)
), defined during the annotation process has been replaced by a cassette containing the gene for kanamycin resistance under the control of a strong promoter (Phage T5 promoter) by homologous recombination. The extremities of the integration cassette, called "recombination footprints" consist of the specific regions which frame the gene to be mutated. The replacement usually covers the totality of the ORF (
2).
However, to avoid altering essential regions such as the RBS (
3)
or the promoters of neighboring genes, we have had to construct a replacement in which the target ORF is truncated by several amino acids. The molecular construction of the cassettes is obtained in detail using PCR (
4)
which recombines the recombination footprints with the gene for antibiotic resistance. After transformation of Acinetobacter baylyi with each of these cassettes, the mutants are selected on a mineral medium supplemented with succinate and kanamycin.
Technique :
Production phase:
Optimization of the transformation protocol and organization of the “pipeline” has permitted us to attain a success rate of over 90%, and a capacity of 400 mutants initiated every two weeks. The complete cycle for the production of a mutant from the transformation step to storage is 4 weeks. This cycle includes control PCRs to check on the replacement of the ORF by the integration cassette and systematic sequencing of the genomic regions of the mutant which are adjacent to the integration cassette.
Results :
Of the 3205 genes annotated in A. baylyi which are candidates for mutation, we have attempted to inactivate 3,167 genes. We produced replacement mutants for 2,594 genes, which represents 81% of the genes of A. baylyi (
de Berardinis
et al., 2008).
Distribution of mutants according to functional category of the genes
This essentiality data set is 88% consistent with the Escherichia coli data set inferred from the Keio mutant collection (Baba et al., 2006) profiled for growth on minimal medium, while 80% of the orthologous genes described as essential in Pseudomonas aeruginosa are also essential in ADP1.
Several strategies were undertaken to investigate ADP1 metabolism by
- searching for discrepancies between our essentiality data and current metabolic knowledge,
- comparing this essentiality data set with those from other organisms,
- systematic phenotyping of the mutant collection on a variety of carbon sources (quinate, 2-3 butanediol, glucose, etc.).
-
This collection provides a new resource for the study of gene function by forward and reverse genetic approaches and constitutes a robust experimental data source for systems biology approaches (Durot et al., en préparation).
To help us in this comparison, a
ADP1 Genome Browser was implemented in order to present data of our mutant collection in their genomic context: KEGG and BioCyc metabolic pathways, SEED subsystems, operon predictions, orthologous genes and essentiality data from several organisms, and phenotyping data from high throughput experiments...
Distribution of the ADP1 mutant collection
Individual mutants have already been distributed and used for several studies as DNA damage (Chakravorty et al., 2008) and analysis of metabolism (Aghaie et al., submitted).
La distribution des mutants est réalisée suivant le protocole ci-dessous qui inclut les données sur les primers utilisés pour réaliser les délétions.
Perspectives :
The systematic gene replacement phase which has now been achieved will be extended by a targeted finishing phase, to attempt to obtain mutants in genes in which the first attempt has failed. Simple repeat of the experiments should suffice for these "technical failures;" new primers in more favorable regions will be selected for some of them. We will also try to repeat the experiments on more enriched media in order to obtain mutants of the genes which are essential on succinate.
An analytic phase will now be undertaken in the collection, in addition to systematic phenotyping in order to study, for example, genes with unknown function which co-localize with genes which participate in the same metabolic pathway.
Glossaire :
(
1)
CDS : Coding DNA Sequence.
(
2)
ORF : Open Reading Frame.
(
3)
RBS : Ribosome Binding Sequence.
(
4)
PCR : Polymerase Chain Reaction.
Strain request for mutants of
Acinetobacter baylyi ADP1
- All strains are free. You will only be charged for the shipping. (The complete collection is not yet available for distribution).
- We will use FedEX international shipping.
-
Please, mail the attached document « request-strain-order »
completed with your personal information to adp1-mutants@genoscope.cns.f . A Material Transfer Agreement (MTA) will be attached on "Confirmation e-mail" sent upon your order.
-
A representative of your research group should print out this MTA and fill in the space.
Please send to the following address. DO NOT send by FAX or e-mail.
Dr. Veronique de Berardinis
Genoscope/CEA
2, rue Gaston Crémieux
CP5706
91057 Evry Cedex
France
- The strains will be dispatch after we receive the MTA.
Primers
data used for target gene deletions and external PCR verifications are available in .xls format