The genetic information carried by the DNA within chromosomes is a precious resource that has to be repeatedly read for cell functions and transmitted to future generations. However, DNA takes a lot of abuse, including copy errors during replication and other accidents occurring due to cellular stress or even just spontaneously. Most often this wear and tear is corrected with surprising precision by the cell's various repair systems, which constantly monitor the DNA to guarantee the integrity of its information.
Telomeres are repetitive sequences of DNA located at the extremities of chromosomes. Their role is to protect genetic information during cell division. The length of these sequences responds to a careful game of maintaining a balance between cellular aging and the risk of uncontrolled cellular proliferation, this latter being a characteristic of cancer. Numerous proteins interact with the telomeres as a part of this balancing game and also to protect their stability. However, a large portion of the multitude of mechanisms involved therein remain mysterious.
In a study published in Nature Communications, researchers from the Telomeres and Chromosome Repair Laboratory of the Genetic Stability, Stem Cells and Radiation Mixed Research Unit (iRCM/CEA-Jacob) and the Nuclear Envelope, Telomeres and DNA Repair team (I2BC/CEA-Joliot) have shed light on one of those telomere protection mechanisms.
In the yeast Saccharomyces cerevisiae, a complex called Mre11-Rad50-Xrs2 (MRX) is involved in the control of telomere length and furthermore plays a fundamental role in DNA double-strand break repair. However, as the natural extremities of chromosomes, telomeres should neither be recognized nor repaired as DNA damage, as doing so would create a risk of inter-telomere fusion or degradation of internal sequences. The MRX complex must thus be specifically inhibited at the telomeres but remain active elsewhere.
In their study, The CEA researchers showed that a protein, Rif2, present at the telomeres and known to block their elongation, is able to bind MRX via a 26-amino-acid motif. That binding can only occur at the telomeres, where the concentration of the motif is adequate. It is also sufficient to keep MRX from binding to DNA, which prevents the risk of fusion at the telomers specifically. Through in vivo genetic approaches guided by in silico structural predictions, the researchers precisely identified the residues of the MRX complex involved in the interaction with Rif2 and thus the residues essential to its inhibition.
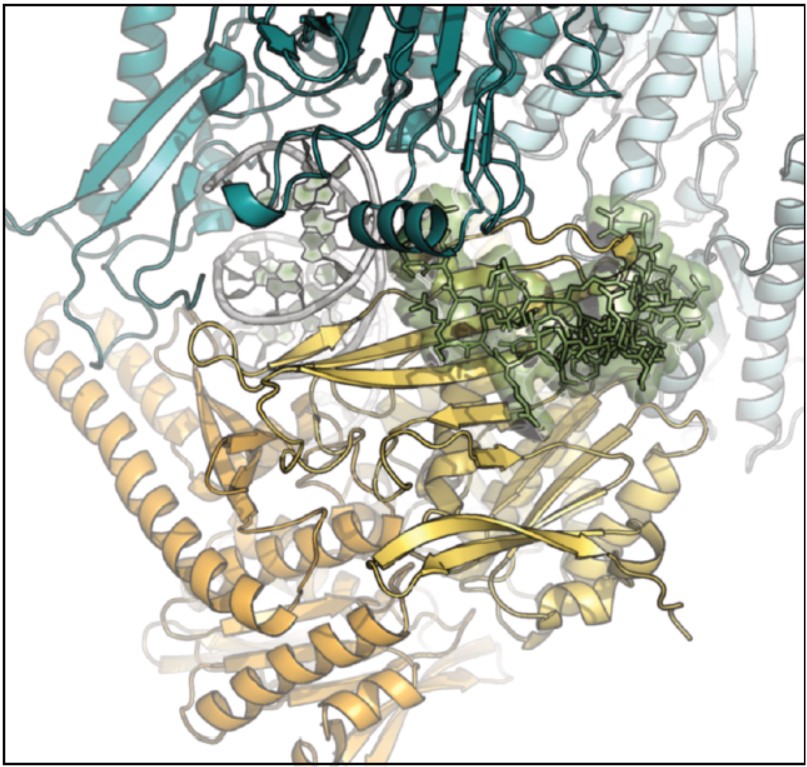
In silico modeling study predicting the most-favorable three-dimensional structures and the amino acids involved in the inter-protein interactions. A small Rif2 motif (center, in green) suffices for binding and binding inhibition to MRX (in blue, orange and yellow).
Modeling studies based on these results enabled a better understanding of this mechanism and the proposal of a model wherein the Rif2-MRX bond favors a MRX three-dimensional structure that is incapable of binding to telomeres and thus spares these latter from uncontrolled actions by the complex.
Already the researchers are looking ahead to the next step, that is, to use this knowledge gained from yeasts, the go-to model for genetic studies, to identify the equivalent mechanism in humans. Indeed, work is already underway to study the possible evolutionary conservation of the mechanism identified in yeast and identify the inhibition mechanism for the MRN complex, the human equivalent of MRX. If this further work is fruitful, the MRN complex could become a therapeutic target to mitigate the uncontrolled cellular proliferation seen in cancer.