While recent advances in spatially resolved “omics” are revolutionizing the exploration of tissue complexity, most available solutions remain focused solely on detecting RNA molecules. However, studies have shown that the half-life of RNA can vary from 40 minutes to 9 hours, meaning that transcriptomic profiling does not directly reflect the transcriptional state of the cell at the time of measurement. Moreover, by definition, transcriptomic analyses can only capture reads from expressed genes, and cannot distinguish between repressed genes and genes maintained in a transcriptionally permissive state, as both appear transcriptionally inactive.
Chromatin immunoprecipitation strategies (using antibodies to capture DNA-associated proteins) to detect histone modifications (proteins that compact DNA) across the genome have become the gold standard for assessing transcriptional regulation at a given time point in samples. More recently, the enzymatic variant known as “Cut&Tag” has increased sensitivity by reducing the number of required cells from several million to just a few thousand, and even down to the single-cell level. This makes it possible to identify histones carrying modifications associated with transcriptional regulation.
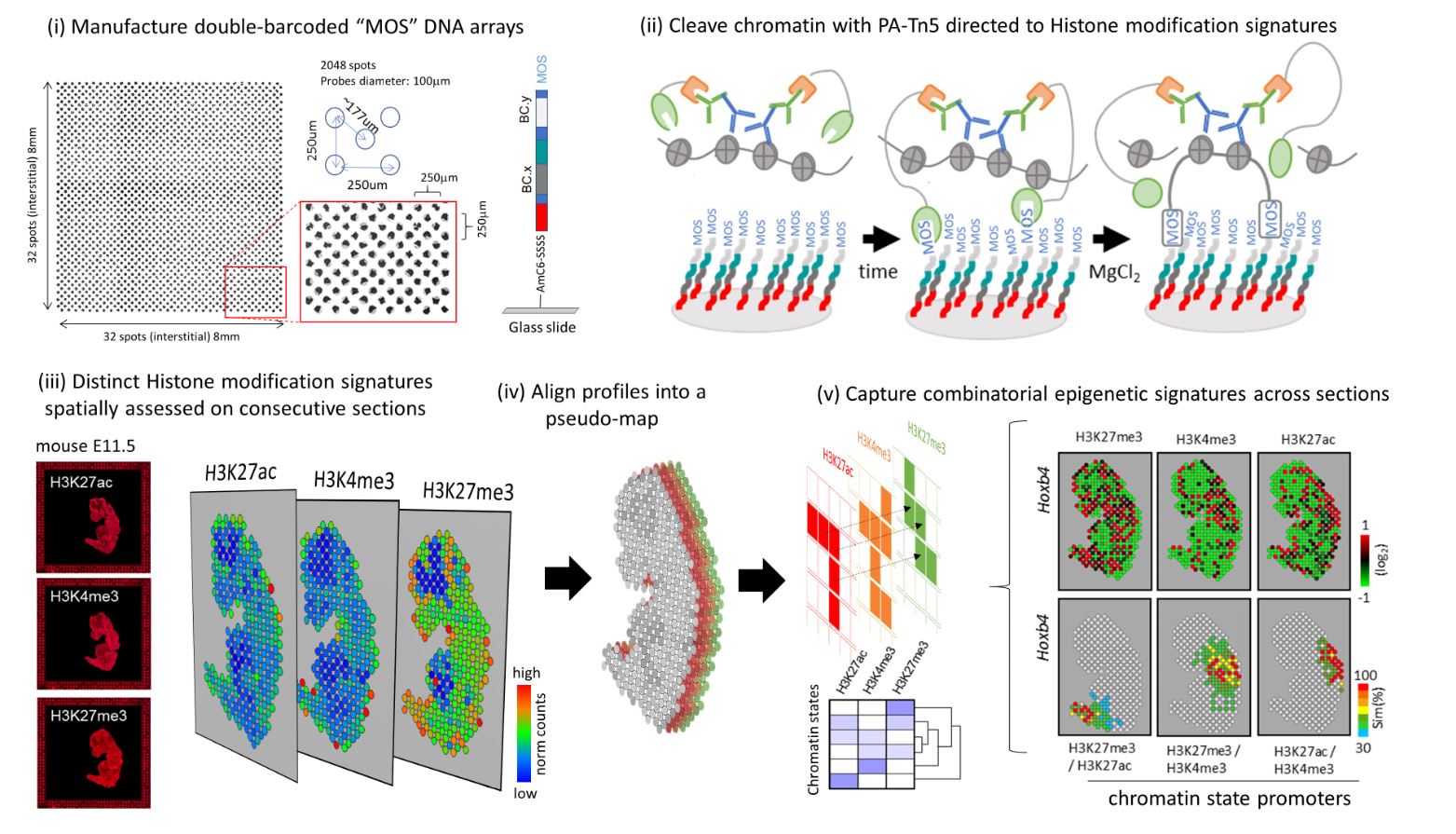
Researchers at Genoscope have developed a new method to spatially identify histone modification signatures using a DNA chip with unique molecular barcodes at a resolution of 100 micrometers. This technology, based on the use of the recombinant protein PA-Tn5, enables chromatin cleavage and immobilization through the creation of a covalent bridge with the chip’s DNA probes. The team validated this approach in adult mouse brains, frozen mouse embryos, and decalcified bone tissue preserved in paraffin blocks (FFPE)—a condition in which RNA is poorly preserved, particularly following decalcification procedures.
An initial technology was developed by a team at Yale University in the United States to map histone modifications on tissue sections using 20-micron microfluidic channels (Deng et al., Science 2022). Although the method developed at Genoscope does not achieve the same resolution, the flexibility of manufacturing DNA chips covering customized surfaces at low cost—thanks to the combinatorial number of required probes—makes it highly effective for analyzing large numbers of tissue sections, including samples of considerable size.
@ M. Mendoza-Parra / CEA
In this study, published in Genome Research, the Genoscope team also emphasized the importance of collecting consecutive tissue sections to acquire distinct histone modification signatures, which can then be combined to reveal chromatin state profiles and compared in a spatial context. This strategy is expected to gain power in the coming years as sequencing costs continue to decrease, enabling the collection of multiple consecutive sections to reconstruct volumetric views of chromatin state signatures that govern tissue architecture.
Contact : mmendoza@genoscope.cns.fr