Group leader
Nadège Jamin
nadege.jamin@cea.fr
Project description
In addition to transversal heterogenity, cell membranes present lateral heterogeneity due non-homogeneously distribution of proteins and lipids within the membrane plane. Transversal and lateral heterogeneity are interdependent. Portions of membranes with distinct composition from the rest of the plasma membrane are named membrane domains. These domains, depending on their composition, have different physical and chemical properties, such as thickness, elasticity, long or short chains saturated lipids, cholesterol, protein crowding, etc... and thus different functionnal roles. Caveolae are « stable » membrane domains enriched in Caveolin-1, Cavins, and specific lipids such as cholesterol. Caveolin-1 is a small monotopic membrane protein found at the plasma membrane in caveolae and non-caveolae domains as caveolin-1 oligomers. It is a key structural protein of caveolae as it possesses the intrinsic properties to curve its membrane. The complex assemblies of caveolin-1 and lipids at the plasma membrane represent a challenge for the understanding of the molecular and functionnal properties of caveolin-1.
In that context, we aim at understanding the relationship between the original amino acid sequence of Caveolin-1 and its properties of oligomerization and shaping the membrane at a molecular level. For that purpose, we have produced and characterized assemblies of caveolin-1 fragments and detergent by high resolution magnetic resonance spectroscopy and modelisation, and we are now pursuing our work using Caveolin-1b , fragments and mutants assemblies with different detergents and lipids using a range of biophysical techniques.
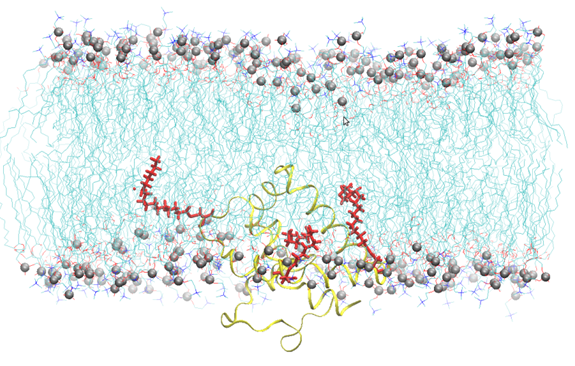
Figure 1: Snapshot of Cav-1(76-178) embedded into a SOPC bilayer after 200 ns of all atoms molecular dynamics simulation. Lipids are represented by blue lines, phosphorus atom of the lipid head groups by silver sphere, Cav-1 (76-178) by a yellow ribbon, the palmitoyl chain by red sticks. Water molecules have been omitted for the sake of clarity.
The overexpression of Caveolin-1β can produce intracellular membrane vesicles called heterologous caveolae (h-caveolae) in the host organisms. We have observed this production in Escherichia coli bacteria and in the Sf21 insect cells, and we propose to produce membrane protein-enriched vesicles using the ability of Caveolin 1β to generate membrane vesicles within the cytoplasm in various hosts. As a proof of concept, we have chosen to co-express with Caveolin-1β the human microsomal glutathione S-transferase (hMGST1) in Escherichia coli and in the Sf21 insect cells. hMGST1 belongs to the membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG) family of integral MPs and catalyze glutathione-dependent modifications of lipophilic substrates present in the lipid membranes. hMGST1 is highly abundant in liver, and displays a noticeable role in xenobiotic detoxification, including some cytotoxic drugs.