Our research goals
Neurodegenerative diseases are largely associated with alterations of cellular metabolism and structure, which may precede neuron death. The magnetic resonance methodology team is developing original methods to non-invasively assess cellular metabolism and structure, in particular in animal models developed within the research unit UMR 9199, using the 7 T and 11.7 T MRI machines in MIRCen. We pursue two ultimate goals: to propose new biomarkers of neurodegenerative diseases, and to better understand physiopathological processes at stake in those diseases.
Figure 1: Quantification of brain metabolites in a volume of the primate brain in vivo at 7 T
Beyond the sole determination of brain metabolite concentrations by proton spectroscopy (Figure 1), our group develops imaging methods based on CEST effect ("Chemical Exchange Saturation Transfer", Figure 2), in order to map with a good spatial resolution the distribution of some endogenous metabolite such as glutamate (which is involved in both energy metabolism and neurotransmission).

Figure 2: Decrease of glutamate concentration observed by CEST imaging in a mouse model of Huntington's disease,
as measured on the 11.7 T scanner. [Adapted from Pépin et al.Neuroimage 2016]
The team is strongly involved in the development of X-nuclei spectroscopy to measure of some important energy metabolism fluxes: carbon-13 (13C) spectroscopy to determine the TCA cycle (VTCA); oxygen-17 (17O) spectroscopy and imaging (Figure 3) to assess the rate of cellular respiration (CMRO2), which is itself coupled to TCA cycle; and phosphorus-31 (31P) spectroscopy to measure the ATP synthesis rate by oxidative phosphorylation (VATP).
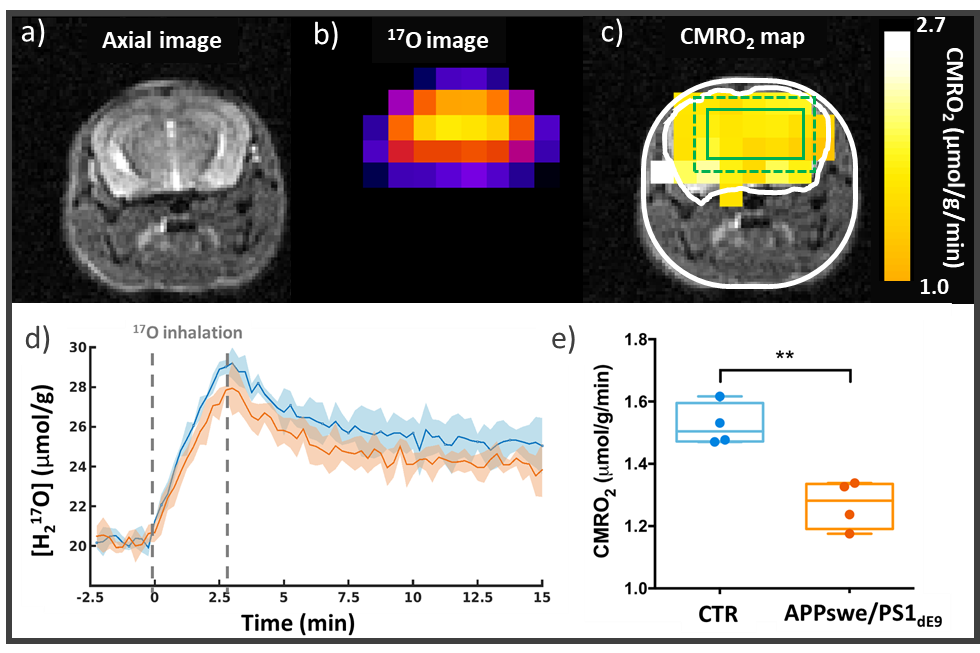
Figure 3: CMRO2 impairment in a mouse model of amyloidosis (APPswe/PS1dE9). a) Anatomical image1H and b) zero echo time (ZTE) 17O images are acquired at 11.7 T. Acquiring a series of 17O-ZTE images before, during, and after inhalation of 17O-labeled oxygen gas (70%) allows the detection of metabolically produced H217O. d) H217O signal time courses in APPswe/PS1dE9 (n=4, orange) and WT mice (n=4, blue) (with SD). (e) Quantification of CMRO2 shows a slower oxygen consumption rate in the APPswe/PS1dE9 mice than in WT (mean±SD).
In parallel, one of our current project deals with the use of CEST imaging of glucose to map cerebral metabolic rate of glucose (CMRglc). An originality of our team is that we combine these different techniques to get an integrated picture of energy metabolism (Figure 4).
Figure 4 Metabolic fluxes of mitochondrial energy synthesis, measured by our group in the primate brain.
The fluxes are in μmol/g/min. Adapted from [Chaumeil et al., PNAS 2009].
We are also looking at the possibility of evaluating the organization of the intracellular medium in an indirect way, by measuring via original diffusion-weighted spectroscopy techniques how this organization constrains the displacement of metabolites. In particular, our group has explored the diffusion of brain metabolites over unprecedented time scales, making it possible to better characterize metabolite compartmentation and the parameters governing metabolite motion. We are also developing new diffusion modeling strategies (in collaboration with Marco Palombo at University College London) to extract quantitative information about the cellular structure from experimental diffusion data. Notably, we have shown that it was possible to differentiate neuronal from astrocytic structure, by studying diffusion of metabolites predominantly in neurons or in astrocytes (Figure 5). We are now investigating the possibility to use diffusion-weighted spectroscopy to assess cerebral lactate distribution between the different compartments (neurons, astrocytes, extracellular space…), which is related to the lactate shuttle. These thematics have been funded by two grants from the European Research Council ("INCELL" and "LactaDiff" projects).
Figure 5: The study of the temporal dependency of metabolite diffusion coefficient at ultra-long diffusion times allows characterizing long-range cellular structure. By looking at metabolites mostly in astrocytes (e.g. myo-inositol) or in neurons (e.g. NAA), it is even possible to differentiate astrocytic from neuronal structure. Taken from [Palombo et al., PNAS 2016]).
Group members
- Julien Valette (CEA researcher): team leader, and leading the diffusion-weighted spectroscopy thematic
- Céline Baligand (CEA researcher): leading the X-nuclei thematic
- Julien Flament (INSERM research officer): leading the CEST thematic
- Eloïse Mougel (post-doctoral fellow): diffusion-weighted spectroscopy sequences
- Rodrigo Lerchundi (post-doctoral fellow): FRET imaging of lactate, enzyme-electrodes
- Amélie Tourais (PhD student): 17O MRI
- Sophie Malaquin (PhD student): diffusion-weighted spectroscopy
- Yohann Mathieu-Daudé (PhD student): CEST imaging of glucose
- Jean-Baptiste Pérot (PhD student): CEST imaging of glutamate
- Mélissa Vincent (PhD student): diffusion-weighted spectroscopy
Past group members
- Jérémy Pépin (PhD student): CEST imaging of glutamate
- Khieu Van Nguyen (post-doctoral fellow): diffusion modeling
- Edwin Hernandez-Garzon (post-doctoral fellow): confocal microscopy, cellular
- Clémence Ligneul (PhD student): diffusion-weighted spectroscopy
- Marco Palombo (post-doctoral fellow): diffusion modeling
- Brice Tiret (PhD student): X-nuclei spectroscopy
- Chloé Najac (PhD student): X-nuclei spectroscopy, diffusion-weighted spectroscopy
- Charlotte Marchadour (PhD student): X-nuclei spectroscopy, diffusion-weighted spectroscopy
Collaborations
- Danish Research Centre for Magnetic Resonance (H. Lundell)
- University of Bordeaux / RMSB (A.-K. Bouzier-Sore)
- University College London (M. Palombo)
- University of Minnesota (P.-G. Henry, M. Marjanska)
- Leiden University (I. Ronen)
- EPFL (C. Cudalbu, R. Gruetter)
- Brain and Spine Institute (F. Branzoli, S.
Lehéricy)
Major grants
ERC: LactaDiff project (2019-2024), INCELL project (2013-2018)
ANR: nrjCEST project (2018-2021); HDeNERGY project (2015-2019)
Some recent publications
Zero Echo Time 17O-MRI Reveals Decreased Cerebral Metabolic Rate of Oxygen Consumption in a Murine Model of Amyloidosis. Baligand C, Barret O, Tourais A, Pérot JB, Thenadey D, Petit F, Liot G, Gaillard MC, Flament J, Dhenain M, Valette J.
Metabolites. 2021;11(5):263. doi: 10.3390/metabo11050263.
Revisiting double diffusion encoding MRS in the mouse brain at 11.7T: Which microstructural features are we sensitive to?Vincent M, Palombo M, Valette J.
Neuroimage 2020;207:116399. doi: 10.1016/j.neuroimage.2019.116399.
Brain Metabolite Diffusion from Ultra-Short to Ultra-Long Time Scales: What Do We Learn, Where Should We Go?Valette J, Ligneul C, Marchadour C, Najac C, Palombo M.
Front Neurosci. 2018;12:2. doi: 10.3389/fnins.2018.00002
Insights into brain microstructure from in vivo DW-MRS.Palombo M, Shemesh N, Ronen I, Valette J.
Neuroimage 2018;182:97-116. doi: 10.1016/j.neuroimage.2017.11.028.
Probing metabolite diffusion at ultra-short time scales in the mouse brain using optimized oscillating gradients and “short” echo time diffusion-weighted MR spectroscopy.Ligneul C, Valette J.
NMR in Biomedicine 2017;30(1). doi: 10.1002/nbm.3671.