A team of researchers from IRCM (CEA-Jacob) and I2BC have shed light on the role of ComFC, a bacterial protein involved in natural transformation, itself a driving mechanism for the propagation of antibiotic resistance genes and virulence factors. Their study, published in Nature Communications, lays pathways for the discovery of novel targets to impede the development of multidrug-resistant bacterial strains.
Antibiotic resistance is not only a major issue in public health but also an accelerating one, with an ever-increasing number of pathogenic bacteria becoming resistant to available antibiotics over the last few years. Natural transformation is a bacterial mechanism for horizontal gene transfer. Through it, bacteria are able to exchange genetic material independently of division: any one bacterium can capture DNA released into the environment by a sister cell—and even other organisms—and incorporate it into its own genome. Particularly, natural transformation plays a major role in the propagation of antibiotic resistance genes and virulence factors among bacterial populations.
During transformation, a bacterial surface protein complex captures free-floating double-strand DNA and transfers it across the membrane (or membranes) and into the cell as a single-strand DNA. Thereafter, a homologous recombination mechanism ensures the integration of the new genetic material into the bacterium's genome. Although these mechanisms were identified several decades ago and the involved proteins are known to be highly conserved among bacteria capable of natural transformation, the underlying molecular mechanisms are poorly understood.
To bring light to that aspect, researchers from the Laboratory of Genetic Instability Research within IRCM's Genetic Stability, Stem Cells and Radiation mixed research unit (CEA-Jacob) teamed with colleagues from I2BC* to characterize ComFC, a key, highly conserved, cytoplasmic transformation protein whose role in the mechanism has, to date, remained mysterious. Their study, published in Nature Communications, was carried out on the species Helicobacter pylori, a model bacterium present in the gut of close to half of the world's population. For many people, this species causes no illness but in others it is the main cause of ulcers and gastric cancers. Because of increasing multidrug-resistance in H. pylori, the World Health Organization has added it to its list of priority pathogens for which new antibiotics or new means to slow antibiotic resistance are needed.
By combining genetic, molecular and cellular approaches, the team showed that ComFC associates with the inner membrane and becomes crucial not only for the transportation of the DNA across that membrane, but also for its handling once in the cytosol. Structural studies of the ComFC protein found a likely phosphoribosyl transferase domain and a zinc finger motif, and genetic & biochemical studies studies showed that both were essential to the protein's function.
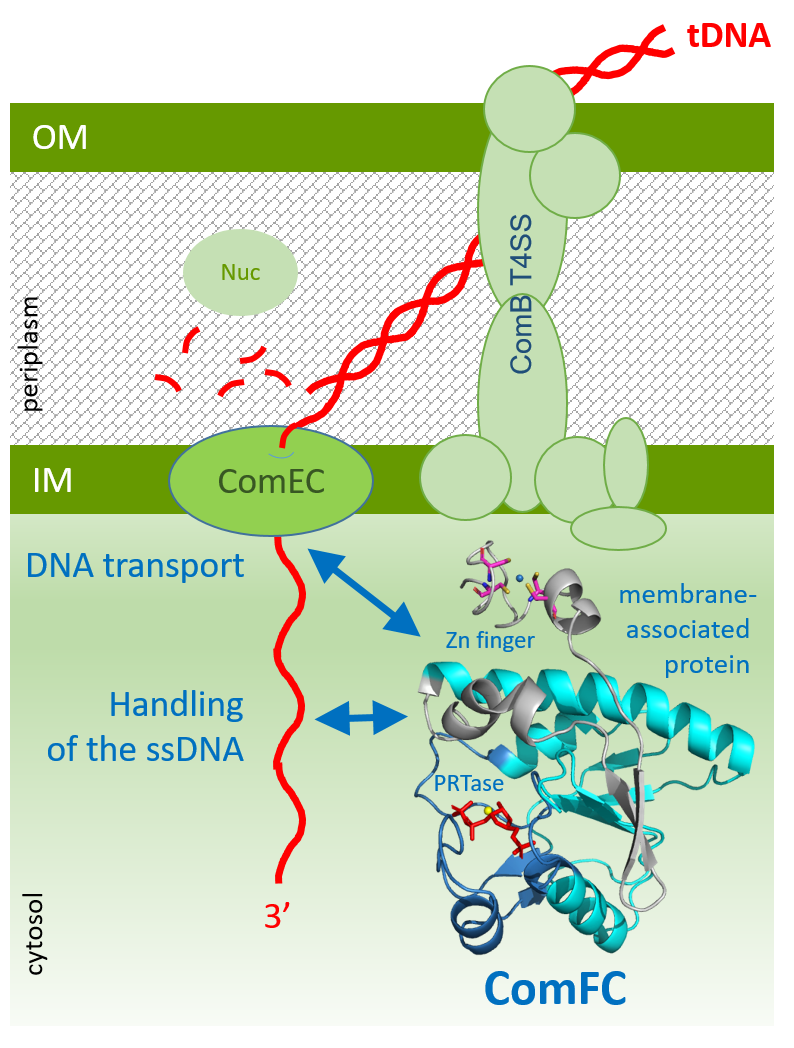
ComFC is a cytoplasmic bacterial protein essential for natural transformation. It is involved in the transport of transforming DNA across the bacterial membrane(s) and the handling of the single-strand DNA delivered to the bacterial cytosol. Structural studies show a zinc-finger motif and a putative phosphoribosyl transferase (PRTases), both necessary for the protein's function. © Sophie Quevillon-Cheruel / I2BC
This work by the IRCM/I2BC team provides a better understanding of the molecular mechanisms driving the horizontal transfer of resistance and virulence in a wide range of pathogenic bacteria. In doing so, it brings new possibilities in the search for novel means to inhibit natural transformation and thus limit virulence and the development of multidrug-resistant bacterial strains.
* Teams Function and architecture of macromolecular assemblies et Molecular assemblies and Genome Integrity, I2BC.