Paramecium, model eukaryotic unicellular organism
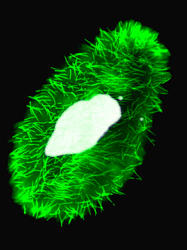
The cilia of the Paramecium, which are colored in green, cover the cell completely and enable the organism to swim and capture its food, which consists of bacteria. To the right of the huge macronucleus colored in light blue, the micronuclei can be distinguished as two little points. (photo J. Beisson)
Well-known to students as a model unicellular organism, Paramecium (Paramecium tetraurelia) is a very large eukaryotic cell (120 micrometers) covered with vibrating cilia. It belongs to the Ciliate phylum (Ciliophora). In the Alveolate clade, the Ciliates form a group related to the unicellular parasites named apicomplexans, which include Plasmodium falciparum, the main causative agent of malaria. Paramecium is an organism which is both unicellular and complex; it is therefore an excellent model for the genetic study of the numerous differentiated functions in multicellular organisms which are absent in simpler eukaryotes such as yeast.
The Ciliates have the fascinating property of having separate germinal and somatic lineages within a single cytoplasmic unit. These cells possess indeed two nuclei. A germinal nucleus (the micronucleus) is responsible for the transmission of genetic information via sexual processes, whereas a somatic nucleus (the macronucleus) ensures expression of this information. At each sexual generation, a new somatic nucleus is produced by programmed rearrangements of the whole genome contained in the germinal nucleus.
Fundamental processes which can be studied in Paramecium.
- These programmed rearrangements of the genome, as well as the massive DNA amplification which produces the somatic nucleus, implicate chromosomal fragmentation, the addition of telomeres and the precise elimination of internal sequences. These processes are similar to somatic rearrangements which are involved in the genesis of diversity in immunoglobulins, and to chromosome rearrangements associated with some cancers in vertebrates. In Paramecium, these rearrangements involve the whole genome and can be easily induced.
- Homology-dependent
gene extinction, a
genome defense mechanism against parasitic nucleic acids, is present in
Paramecium, and has been conserved during the evolution of eukaryotes. This
mechanism, also known as RNA interference, may be experimentally triggered by
the introduction of a transgene or double-stranded RNA into the cell. It acts
at the post-transcriptional level and leads to the degradation of all
homologous messenger RNAs in the cell. RNA interference is also implicated in
the development of the somatic nucleus of the Paramecium, a fact which can
explain the non-Mendelian transmission of certain traits in this ciliate.
Paramecia are privileged organisms for the study of such epigenetic phenomena,
which are easily accessible for experimentation in these cells. The biologic
role of epigenetic phenomena, in the development of multi-cellular organisms
for instance, is not yet fully understood.
- Paramecium is also capable of regulated exocytosis of secretory vesicules (trichocysts) in response to external stimulation, like that which occurs in animals during the secretion of hormones or neuromediators. This regulated secretion implies a membrane fusion stage triggered by a transmembrane signaling cascade. In Paramecium it functions as a defense against predators, but is not essential under culture conditions in the laboratory. Consequently, Paramecium provides one of the rare models in which regulated secretion, and especially the last stage of membrane fusion during exocytosis, can be dissected using a genetic approach.
- The membrane of Paramecium is excitable. As in neurons and muscle cells, thismembrane excitability is governed by the activity of ion channels and membrane receptors. In Paramecium, their function controls the speed and direction of the beating of the cilia of the cell. The activity of these channels can therefore be easily observed by watching the swimming behavior. It is in Paramecium that the first mutants for ion channels were isolated at the end of the sixties, as well as the first mutants for calmodulin (an ubiquitous protein which regulates many cell processes in response to changes in calcium concentration).
- Paramecium possesses basal bodies, organelles which are homologous to the centrioles which are present in multicellular organisms. These complex protein structures have the remarkable property of duplicating themselves at each cell division by a mechanism which has not yet been elucidated. The duplication of the basal bodies plays a key role in cell division and organization, as in the morphogenesis of the Paramecium. The genetic dissection of this phenomenon, for which Paramecium constitutes an extremely advantageous model, has already led to the discovery of new proteins.
Two essential tools available in Paramecium: effective genetics...

The use of antibodies directed against tubuline reveal the Paramecium basal bodies, located at the base of the cilia. The macronucleus and the micronuclei (the two little points close to the macronucleus on its right side) appear again in blue. (Photo F. Ruiz)
More than 50 years of classical genetics experiments have led to the accumulation of almost 200 Mendelian mutations of Paramecium, affecting very diverse cellular processes (i.e. morphogenesis, regulated secretion, cell cycle, antigenic variation, sex determination and expression of the mating type, and rearrangements of the genome). Indeed, Paramecium is very well-suited to genetic analysis because of its two modes of sexual reproduction, autogamy and conjugation. Autogamy is a process of self-fertilization which renders the genome of the zygote completely homozygous in one generation. The stored strains are therefore just as easy to manipulate as haploid organisms. Conjugation is a process of reciprocal fertilization which produces two new zygotic nuclei which are identical in the two partners, which makes it possible to identify the traits with Mendelian heredity very easily, and to distinguish traits which are maternally inherited. The genes identified by mutation can be cloned by functional complementation.
...and functional genetics using RNA interference
Gene silencing, which can be provoked by the introduction of transgenes, provides a powerful tool for the functional analysis of genomes. However, the ideal tool in Paramecium is RNA interference, which can be obtained with remarkable efficiency in this organism by ingestion of bacteria which produce double-stranded RNA. This method of "feeding," which was originally developed for the nematode Caenorhabditis elegans, makes it possible to envisage large scale functional analysis of the ORFs that are identified.
Paramecium and its "two" genomes
The nuclei of Paramecium, the micronucleus and the macronucleus, differ in both structure and function. The diploid micronucleus, which is present in two copies in the P. tetraurelia species, represents the germ line and is completely silent in terms of transcription. This is the nucleus which undergoes meiosis and fertilization during sexual events (conjugation between competent cells or autogamy in a single cell).
The macronucleus, which is highly polyploid (about 1000n), represents the somatic line and is the site of transcription. Both the macronucleus and the micronucleus are derived from copies of the zygotic nucleus. The programmed development of the macronucleus includes DNA amplification by a factor of about 250, precise elimination of short internal sequences called IES and imprecise elimination of regions which are rich in transposons and repeated sequences and are probably heterochromatic. These events causes fragmentation of chromosomes. The extremities created in this way are repaired by the addition of telomeres.
Comparative properties of the micronucleus and macronucleus
| Micronucleus | Macronucleus |
| Ploidy | 2 n | 1000 n |
| Genome size | 100-120 megabases | 80-90% of the micronuclear complexity |
| Number of chromosomes | 50 | 350 |
| Chromosome size | 2 megabases | 300 (50-1000) kilobases |
The majority of the heterochromatin is eliminated during development of the macronucleus, which is therefore essentially euchromatic and devoid of repeat sequences and microsatellites. This represents a real advantage for a first genome sequencing project, because the presence of repeat sequences causes technical problems, notably during the genome assembly stage.
Because of this elimination of repeated sequences, the macronuclear genome of Paramecium is very "compact": it is estimated that its coding fraction is over 70% (in the human genome, it is of the order of 1%!). The introns are uniformly small (from 18 to 35 bases) and the intergenic regions are generally less than 50 100 bases, and may be only a few bases long (a minimum of 9 bases has been observed to date). Its remarkably compact nature makes the macronucleus the material of choice for the inventory of Paramecium genes.
This inventory will also benefit from the sequencing of the genome of Tetrahymena thermophila, which is another Ciliate, evolutionarily distant from Paramecium by over 100 million years. Tetrahymena is also being studied by a large community of scientists.
History of the Paramecium sequencing project
- August 1999: Following an initiative of the "Genetics of cellular dynamics in Paramecium" Group at the CNRS Molecular Genetics Center in Gif-sur-Yvette, a consortium of 11 European and North American laboratories was formed to autofinance a pilot project for the random sequencing of the genome of Paramecium. This financing produced the sequences of 3000 plasmid insert ends from a macronuclear DNA library. The annotated sequences were deposited in the GSS division of public databases in November 2000 (P. Dessen et al. 2001, Trends in Genetics 17: 306-308; L. Sperling et al. 2002, Euk. Cell 1:341-352).
- October 2000: Meeting of the European members of this Consortium in Warsaw to define the next stage after the pilot project. It was decided to isolate and sequence one chromosome of the macronucleus which was one megabase long. This project was begun at the end of 2001, and is almost finished in mid-2003.
- July 2002: The CNRS supported the European scientific community by creating an"European Research Group" (Groupement de recherche européen, GRDE) for the Paramecium project, that is, a structure with financing for meetings and exchanges between groups. A first meeting was held in Dourdan in October 2002.
- October 2002: the CNRS provided support for the Paramecium genome project. It allocated a budget of 100,000 euros, within the framework of its large-scale sequencing program, in order to begin the sequencing of the DNA of the Paramecium macronucleus at Genoscope.
- April 2003: The Scientific Council of Genoscope formally approved the total sequencing of the Paramecium macronucleus using Genoscope's own funds.